| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,370,643 – 18,370,733 |
| Length | 90 |
| Max. P | 0.824059 |

| Location | 18,370,643 – 18,370,733 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.78 |
| Shannon entropy | 0.37200 |
| G+C content | 0.32717 |
| Mean single sequence MFE | -18.62 |
| Consensus MFE | -12.63 |
| Energy contribution | -12.88 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.824059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
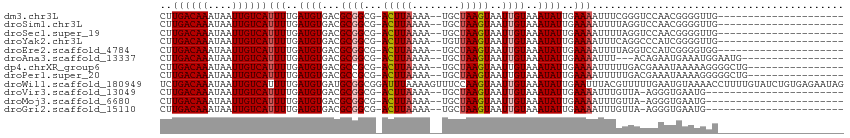
>dm3.chr3L 18370643 90 + 24543557 CUUGACAAAUAAUUGUCAUUUUGAUGUGACGCGGCG-ACUUAAAA--UGCUAAGUAAUUGUAAAUAUUGAAAAUUUCGGGUCCAACGGGGUUG--------------------- ..((((((....)))))).((..((((...((((..-(((((...--...)))))..))))..))))..))(((..((.......))..))).--------------------- ( -19.60, z-score = -1.34, R) >droSim1.chr3L 17705922 90 + 22553184 CUUGACAAAUAAUUGUCAUUUUGAUGUGACGCGGCG-ACUUAAAA--UGCUAAGUAAUUGUAAAUAUUGAAAAUUUUAGGUCCAACGGGGUUG--------------------- ..((((((....))))))...........(.((..(-((((((((--(....((((........))))....))))))))))...)).)....--------------------- ( -18.30, z-score = -1.24, R) >droSec1.super_19 366503 90 + 1257711 CUUGACAAAUAAUUGUCAUUUUGAUGUGACGCGGCG-ACUUAAAA--UGCUAAGUAAUUGUAAAUAUUGAAAAUUUUAGGUCCAACGGGGUUG--------------------- ..((((((....))))))...........(.((..(-((((((((--(....((((........))))....))))))))))...)).)....--------------------- ( -18.30, z-score = -1.24, R) >droYak2.chr3L 8525863 90 - 24197627 CUUGACAAAUAAUUGUCAUUUUGAUGUGACGCGGCG-ACUUAAAA--UGUUAAGUAAUUGUAAAUAUUGAAAAUUUCAGGCCCAUCGGGGUUG--------------------- ..((((((....))))))(((..((((...((((..-((((((..--..))))))..))))..))))..)))......(((((....))))).--------------------- ( -21.80, z-score = -2.02, R) >droEre2.scaffold_4784 10241004 90 - 25762168 CUUGACAAAUAAUUGUCAUUUUGAUGUGACGCGGCG-ACUUAAAA--UGCUAAGUAAUUGUAAAUAUUGAAAAUUUUAGGUCCAUCGGGGUGG--------------------- ...(((((....)))))(((((((((.(((((((..-(((((...--...)))))..)))).....((((.....))))))))))))))))..--------------------- ( -18.80, z-score = -1.24, R) >droAna3.scaffold_13337 4270728 91 - 23293914 CUUGACAAAUAAUUGUCAUUUUGAUGUGACGCGGCG-ACUUAAAA--UGCUAAGUAAUUGUAAAUAUUGAAAAUUU---ACAGAAUGAAAUGGAAUG----------------- ..((((((....))))))(((..((((...((((..-(((((...--...)))))..))))..))))..)))....---..................----------------- ( -15.40, z-score = -1.42, R) >dp4.chrXR_group6 11265325 95 + 13314419 CUUGACAAAUAAUUGUCAUUUUGAUGUGACGCCGCG-ACUUAAAA--UGCUAAGUAAUUGUAAAUAUUGAAAAUUUUUGACGAAAUAAAAAGGGGCUG---------------- ..............(((((......)))))(((.(.-(((((...--...)))))..((((......(....)......))))........).)))..---------------- ( -15.10, z-score = -0.68, R) >droPer1.super_20 1165955 95 + 1872136 CUUGACAAAUAAUUGUCAUUUUGAUGUGACGCCGCG-ACUUAAAA--UGCUAAGUAAUUGUAAAUAUUGAAAAUUUUUGACGAAAUAAAAGGGGGCUG---------------- ..............(((((......)))))(((.(.-(((((...--...)))))..((((......(....)......))))........).)))..---------------- ( -14.90, z-score = -0.38, R) >droWil1.scaffold_180949 518755 114 + 6375548 UCUGACAAAUAAUUGUCAUUUUGAUGUGAUGCGGCGGAUUUAAAAGUUUCCAAGUAAUUGUAAAUAUUGAAUUUACGUUUUUGAAUGUAAAACCUUUUGUAUCUGUGAGAAUAG ...(((((....)))))(((((.(((.(((((((.((.(((((..((((.(((....))).)))).)))))((((((((....)))))))).))..)))))))))).))))).. ( -21.60, z-score = -1.22, R) >droVir3.scaffold_13049 21564421 87 + 25233164 CUUGACAAAUAAUUGUCAUUUUGAUGUGACGCGGCG-ACUUAAAA--UGCUAAGUAAUUGUAAAUAUUGAAAAUUUGUUA-AGGGUGAAUG----------------------- ((((((((((....(((((......)))))((((..-(((((...--...)))))..))))...........))))))))-))........----------------------- ( -19.90, z-score = -2.99, R) >droMoj3.scaffold_6680 16027128 87 + 24764193 CUUGACAAAUAAUUGUCAUUUUGAUGUGACGCGGCG-ACUUAAAA--UGCUAAGUAAUUGUAAAUAUUGAAAAUUUGUUA-AGGGUGAAUG----------------------- ((((((((((....(((((......)))))((((..-(((((...--...)))))..))))...........))))))))-))........----------------------- ( -19.90, z-score = -2.99, R) >droGri2.scaffold_15110 2525800 87 - 24565398 CUUGACAAAUAAUUGUCAUUUUGAUGUGACGCGGCG-ACUUAAAA--UGCUAAGUAAUUGUAAAUAUUGAAAAUUUGUUA-AGGGUGAAUG----------------------- ((((((((((....(((((......)))))((((..-(((((...--...)))))..))))...........))))))))-))........----------------------- ( -19.90, z-score = -2.99, R) >consensus CUUGACAAAUAAUUGUCAUUUUGAUGUGACGCGGCG_ACUUAAAA__UGCUAAGUAAUUGUAAAUAUUGAAAAUUUUUGAUCCAAUGAAGUGG_____________________ ..((((((....))))))(((..((((...((((...(((((........)))))..))))..))))..))).......................................... (-12.63 = -12.88 + 0.26)


| Location | 18,370,643 – 18,370,733 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.78 |
| Shannon entropy | 0.37200 |
| G+C content | 0.32717 |
| Mean single sequence MFE | -11.52 |
| Consensus MFE | -7.12 |
| Energy contribution | -7.05 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.706127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18370643 90 - 24543557 ---------------------CAACCCCGUUGGACCCGAAAUUUUCAAUAUUUACAAUUACUUAGCA--UUUUAAGU-CGCCGCGUCACAUCAAAAUGACAAUUAUUUGUCAAG ---------------------.......(.(((((.((((((.......))))......((((((..--..))))))-...)).))).)).)....((((((....)))))).. ( -11.60, z-score = -0.86, R) >droSim1.chr3L 17705922 90 - 22553184 ---------------------CAACCCCGUUGGACCUAAAAUUUUCAAUAUUUACAAUUACUUAGCA--UUUUAAGU-CGCCGCGUCACAUCAAAAUGACAAUUAUUUGUCAAG ---------------------......(((.((((.((((((......((........))......)--))))).))-).).)))...........((((((....)))))).. ( -11.30, z-score = -1.16, R) >droSec1.super_19 366503 90 - 1257711 ---------------------CAACCCCGUUGGACCUAAAAUUUUCAAUAUUUACAAUUACUUAGCA--UUUUAAGU-CGCCGCGUCACAUCAAAAUGACAAUUAUUUGUCAAG ---------------------......(((.((((.((((((......((........))......)--))))).))-).).)))...........((((((....)))))).. ( -11.30, z-score = -1.16, R) >droYak2.chr3L 8525863 90 + 24197627 ---------------------CAACCCCGAUGGGCCUGAAAUUUUCAAUAUUUACAAUUACUUAACA--UUUUAAGU-CGCCGCGUCACAUCAAAAUGACAAUUAUUUGUCAAG ---------------------.......(((((((.(((.....)))............((((((..--..))))))-.))).)))).........((((((....)))))).. ( -17.10, z-score = -2.58, R) >droEre2.scaffold_4784 10241004 90 + 25762168 ---------------------CCACCCCGAUGGACCUAAAAUUUUCAAUAUUUACAAUUACUUAGCA--UUUUAAGU-CGCCGCGUCACAUCAAAAUGACAAUUAUUUGUCAAG ---------------------.......(((((((...((((.......))))......((((((..--..))))))-......))).))))....((((((....)))))).. ( -13.80, z-score = -2.29, R) >droAna3.scaffold_13337 4270728 91 + 23293914 -----------------CAUUCCAUUUCAUUCUGU---AAAUUUUCAAUAUUUACAAUUACUUAGCA--UUUUAAGU-CGCCGCGUCACAUCAAAAUGACAAUUAUUUGUCAAG -----------------...............(((---((((.......)))))))...((((((..--..))))))-..................((((((....)))))).. ( -11.10, z-score = -2.29, R) >dp4.chrXR_group6 11265325 95 - 13314419 ----------------CAGCCCCUUUUUAUUUCGUCAAAAAUUUUCAAUAUUUACAAUUACUUAGCA--UUUUAAGU-CGCGGCGUCACAUCAAAAUGACAAUUAUUUGUCAAG ----------------..(((..(((((........)))))..................((((((..--..))))))-...)))............((((((....)))))).. ( -10.90, z-score = -1.23, R) >droPer1.super_20 1165955 95 - 1872136 ----------------CAGCCCCCUUUUAUUUCGUCAAAAAUUUUCAAUAUUUACAAUUACUUAGCA--UUUUAAGU-CGCGGCGUCACAUCAAAAUGACAAUUAUUUGUCAAG ----------------..(((.................((((.......))))......((((((..--..))))))-...)))............((((((....)))))).. ( -10.70, z-score = -1.21, R) >droWil1.scaffold_180949 518755 114 - 6375548 CUAUUCUCACAGAUACAAAAGGUUUUACAUUCAAAAACGUAAAUUCAAUAUUUACAAUUACUUGGAAACUUUUAAAUCCGCCGCAUCACAUCAAAAUGACAAUUAUUUGUCAGA ...........(((..((((((((((.......)))))((((((.....)))))).............)))))..)))..................((((((....)))))).. ( -14.30, z-score = -1.47, R) >droVir3.scaffold_13049 21564421 87 - 25233164 -----------------------CAUUCACCCU-UAACAAAUUUUCAAUAUUUACAAUUACUUAGCA--UUUUAAGU-CGCCGCGUCACAUCAAAAUGACAAUUAUUUGUCAAG -----------------------........((-(.((((((.................((((((..--..))))))-......((((........))))....)))))).))) ( -8.70, z-score = -1.93, R) >droMoj3.scaffold_6680 16027128 87 - 24764193 -----------------------CAUUCACCCU-UAACAAAUUUUCAAUAUUUACAAUUACUUAGCA--UUUUAAGU-CGCCGCGUCACAUCAAAAUGACAAUUAUUUGUCAAG -----------------------........((-(.((((((.................((((((..--..))))))-......((((........))))....)))))).))) ( -8.70, z-score = -1.93, R) >droGri2.scaffold_15110 2525800 87 + 24565398 -----------------------CAUUCACCCU-UAACAAAUUUUCAAUAUUUACAAUUACUUAGCA--UUUUAAGU-CGCCGCGUCACAUCAAAAUGACAAUUAUUUGUCAAG -----------------------........((-(.((((((.................((((((..--..))))))-......((((........))))....)))))).))) ( -8.70, z-score = -1.93, R) >consensus _____________________CAACUUCAUUCGAUCAAAAAUUUUCAAUAUUUACAAUUACUUAGCA__UUUUAAGU_CGCCGCGUCACAUCAAAAUGACAAUUAUUUGUCAAG ...........................................................((((((......))))))...................((((((....)))))).. ( -7.12 = -7.05 + -0.07)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:41 2011