| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,362,326 – 18,362,422 |
| Length | 96 |
| Max. P | 0.923524 |

| Location | 18,362,326 – 18,362,417 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 65.32 |
| Shannon entropy | 0.60184 |
| G+C content | 0.43967 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -9.90 |
| Energy contribution | -11.07 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920053 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
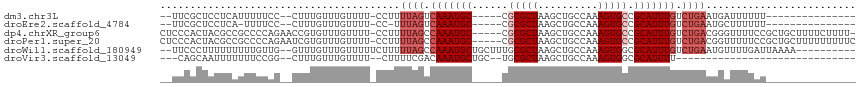
>dm3.chr3L 18362326 91 - 24543557 --UUCGCUCCUCAUUUUUCC--CUUUGUUUGUUUU-CCUUUUAGUCAAAUGC-----CGCGCUAAGCUGCCAAAGUGCCGCAUUUGUCUGAAUGAUUUUUU--------------- --..................--............(-(..(((((.(((((((-----.(((((..........))))).))))))).))))).))......--------------- ( -20.10, z-score = -3.14, R) >droEre2.scaffold_4784 10232763 89 + 25762168 --UUCGCUCCUCA-UUUUCC--CUUUGUUUGUUUU-CC-UUUAGUCAAAUGC-----CGCGCUAAGCUGCCAAAGUGCCGCAUUUGUCUGAAUGCUUUUUU--------------- --...........-......--.............-..-(((((.(((((((-----.(((((..........))))).))))))).))))).........--------------- ( -19.70, z-score = -2.94, R) >dp4.chrXR_group6 11255746 109 - 13314419 CUCCCACUACGCCGCCCCAGAACCGUGUUUGUUUU-CCUUUUAGCCAAAUGC-----CGCGCUAAGCUGCCAAAGUGCCGCAUUUGUCUGACGGGUUUUCCGCUGCUUUUCUUUU- ..........((.((..((((......))))....-(((.((((.(((((((-----.(((((..........))))).))))))).)))).)))......)).)).........- ( -26.10, z-score = -0.98, R) >droPer1.super_20 1156255 110 - 1872136 CUCCCACUACGCCGCCCCAGAAUCGUGUUUGUUUU-CCUUUUAGCCAAAUGC-----CGCGCUAAGCUGCCAAAGUGCCGCAUUUGUCUGACGGUUUUUCCGCUGCUUUUUUUUUC ..........((.((...((((((((.........-.....(((.(((((((-----.(((((..........))))).))))))).)))))))))))...)).)).......... ( -25.43, z-score = -1.76, R) >droWil1.scaffold_180949 5847532 102 + 6375548 --UUCCCUUUUUUUUUGUUG--GUUUGUUUGUUUUUCUUUUUAGCCAAAUGCUGCUUUGCGCUAAGCUGCCAAAGUGGCGCAUUUGUCUGAAUGUUUUGAUUAAAA---------- --...............(((--(((.....(.....).....))))))..((..(((((.((......)))))))..))((((((....))))))...........---------- ( -21.90, z-score = -1.22, R) >droVir3.scaffold_13049 3682945 77 + 25233164 ---CAGCAAUUUUUUUCCGG--CUUUGUUUGUUUU--CUUUUCGACAAAUGCUGC--UGCGCUAAGCUGCCAAAGUGGCGCAUUUU------------------------------ ---...............((--(..((((((((..--......))))))))..))--)(((((..(((.....)))))))).....------------------------------ ( -17.70, z-score = 0.23, R) >consensus __UCCGCUACUCAUUUCCAG__CUUUGUUUGUUUU_CCUUUUAGCCAAAUGC_____CGCGCUAAGCUGCCAAAGUGCCGCAUUUGUCUGAAUGUUUUUCU_______________ ........................................((((.(((((((......(((((..........))))).))))))).))))......................... ( -9.90 = -11.07 + 1.17)

| Location | 18,362,326 – 18,362,422 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.88 |
| Shannon entropy | 0.36427 |
| G+C content | 0.47938 |
| Mean single sequence MFE | -23.91 |
| Consensus MFE | -19.70 |
| Energy contribution | -20.70 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923524 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 18362326 96 - 24543557 CUGCUUUCGCUCCUCAUUUUUCC----CUUUGUUUGUUUUCCUUUUAGUCAAAUGCCGCGCUAAGCUGCCAAAGUGCCGCAUUUGUCUGAAUGAUUUUUU----------- ..((....)).............----............((..(((((.(((((((.(((((..........))))).))))))).))))).))......----------- ( -20.50, z-score = -2.65, R) >droEre2.scaffold_4784 10232763 94 + 25762168 CGGCUUUCGCUCCUCA-UUUUCC----CUUUGUUUGUUUUCC-UUUAGUCAAAUGCCGCGCUAAGCUGCCAAAGUGCCGCAUUUGUCUGAAUGCUUUUUU----------- .(((....))).....-......----...............-(((((.(((((((.(((((..........))))).))))))).))))).........----------- ( -20.90, z-score = -1.81, R) >dp4.chrXR_group6 11255749 111 - 13314419 GGACUCUCCCACUACGCCGCCCCAGAACCGUGUUUGUUUUCCUUUUAGCCAAAUGCCGCGCUAAGCUGCCAAAGUGCCGCAUUUGUCUGACGGGUUUUCCGCUGCUUUUCU (((....(((((.((((.(........).))))..)).......((((.(((((((.(((((..........))))).))))))).)))).)))...)))........... ( -27.70, z-score = -0.63, R) >droPer1.super_20 1156259 111 - 1872136 GGACUCUCCCACUACGCCGCCCCAGAAUCGUGUUUGUUUUCCUUUUAGCCAAAUGCCGCGCUAAGCUGCCAAAGUGCCGCAUUUGUCUGACGGUUUUUCCGCUGCUUUUUU ((......)).....((.((...((((((((..............(((.(((((((.(((((..........))))).))))))).)))))))))))...)).))...... ( -26.53, z-score = -1.00, R) >consensus CGACUCUCCCACCACACCGCCCC____CCGUGUUUGUUUUCCUUUUAGCCAAAUGCCGCGCUAAGCUGCCAAAGUGCCGCAUUUGUCUGAAGGAUUUUCC___________ .........................................(((((((.(((((((.(((((..........))))).))))))).))))))).................. (-19.70 = -20.70 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:40 2011