| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,640,947 – 5,641,049 |
| Length | 102 |
| Max. P | 0.938171 |

| Location | 5,640,947 – 5,641,049 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.53 |
| Shannon entropy | 0.43855 |
| G+C content | 0.38526 |
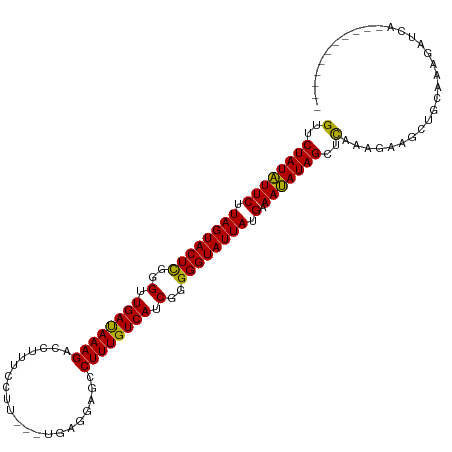
| Mean single sequence MFE | -25.35 |
| Consensus MFE | -21.17 |
| Energy contribution | -20.26 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
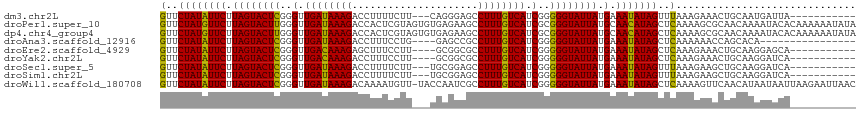
>dm3.chr2L 5640947 102 + 23011544 GUUCUAUAUUCUUAGUACUCGGGUUGAUAAAGACCUUUUCUU---CAGGGAGCCUUUGUCAUCGGGGGUAUUAUGAAAUAUAGUUUAAAGAAACUGCAAUGAUUA----------- ...((((((((.((((((((..(.((((((((.((((.....---.))))...)))))))).)..)))))))).).)))))))......................----------- ( -23.40, z-score = -1.04, R) >droPer1.super_10 1231158 116 - 3432795 GUUCUAUGUUCUUAGUACUUGGGUUGAUAAAGACCACUCGUAGUGUGAGAAGCCUUUGUCAUCGCGGGUAUUAUGCAACAUAGCUCAAAAGCGCAACAAAAUACACAAAAAAUAUA (..((((((((.(((((((((.(.((((((((.(..((((.....))))..).)))))))).).))))))))).).)))))))..).............................. ( -29.90, z-score = -2.37, R) >dp4.chr4_group4 3404248 116 - 6586962 GUUCUAUGUUCUUAGUACUUGGGUUGAUAAAGACCACUCGUAGUGUGAGAAGCCUUUGUCAUCGCGGGUAUUAUGCAACAUAGCUCAAAAGCGCAACAAAAUACACAAAAAAUAUA (..((((((((.(((((((((.(.((((((((.(..((((.....))))..).)))))))).).))))))))).).)))))))..).............................. ( -29.90, z-score = -2.37, R) >droAna3.scaffold_12916 11084724 96 + 16180835 GUUCUAUAUUCUUAGUACUCGGGUUGAUAAAGACCUUUCCUG----GAGCCGCCUUUGUCAUCGGGGGUAUUAUGAAAUAUAGCUCAAAAAACCAGCACA---------------- (..((((((((.((((((((..(.((((((((..(((.....----)))....)))))))).)..)))))))).).)))))))..)..............---------------- ( -21.50, z-score = -0.36, R) >droEre2.scaffold_4929 5729422 101 + 26641161 GUUCUAUAUUCUUAGUACUCGGGUUGACAAAGAGCUUUCCUU----GCGGCGCCUUUGUCAUCGGGGGUAUUAUGAAAUAUAGCUCAAAGAAACUGCAAGGAGCA----------- (((((...(((.((((((((..(.((((((((.(((......----..)))..)))))))).)..)))))))).))).....((...........))..))))).----------- ( -29.40, z-score = -1.73, R) >droYak2.chr2L 8772094 101 - 22324452 GUUCUAUAUUCUUAGUACUCGGGUUGACAAAGACCUUUCCUU----GCGGCGCCUUUGUCAUCGGGGGUAUUAUGAAAUAUAGCUCAAAGAAACUGCAAGGAUCA----------- ((.(((......))).))..(((((......))))).(((((----((((.(((((((....)))))))....(((........)))......)))))))))...----------- ( -26.60, z-score = -1.36, R) >droSec1.super_5 3718688 102 + 5866729 GUUCUAUAUUCUUAGUACUCGGGUUGAUAAAGACCUUUUCUU---UGCGGAGCCUUUGUCAUCGGGGGUAUUAUGAAAUAUAGUUUAAAGAAGCUGCAAGGAUCA----------- ((.(((......))).))..(((((......)))))..((((---(((((.(((((((....)))))))((((((...)))))).........)))))))))...----------- ( -23.30, z-score = -0.46, R) >droSim1.chr2L 5434033 102 + 22036055 GUUCUAUAUUCUUAGUACUCGGGUUGAUAAAGACCUUUUCUU---UGCGGAGCCUUUGUCAUCGGGGGUAUUAUGAAAUAUAGUUUAAAGAAGCUGCAAGGAUCA----------- ((.(((......))).))..(((((......)))))..((((---(((((.(((((((....)))))))((((((...)))))).........)))))))))...----------- ( -23.30, z-score = -0.46, R) >droWil1.scaffold_180708 3937248 115 - 12563649 GUUCUAUAUUCUUAGUACUCGGGUUGAUAAAGACAAAAUGUU-UACCAAUCGCCUUUGUCAUCGGGGGUAUUAUGAAAUAUAGCUCAAAAGUUCAACAUAAUAAUUAAGAAUUAAC .......(((((((((.(((.(((.(((((((..........-..........)))))))))).))).(((((((......((((....))))...)))))))))))))))).... ( -20.85, z-score = -1.21, R) >consensus GUUCUAUAUUCUUAGUACUCGGGUUGAUAAAGACCUUUCCUU___UGAGGAGCCUUUGUCAUCGGGGGUAUUAUGAAAUAUAGCUCAAAGAAGCUGCAAAGAUCA___________ (..((((((((.((((((((..(.((((((((.....................)))))))).)..)))))))).).)))))))..).............................. (-21.17 = -20.26 + -0.91)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:33 2011