| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,316,612 – 18,316,757 |
| Length | 145 |
| Max. P | 0.701568 |

| Location | 18,316,612 – 18,316,757 |
|---|---|
| Length | 145 |
| Sequences | 4 |
| Columns | 163 |
| Reading direction | forward |
| Mean pairwise identity | 64.99 |
| Shannon entropy | 0.55937 |
| G+C content | 0.29915 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -13.46 |
| Energy contribution | -13.78 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
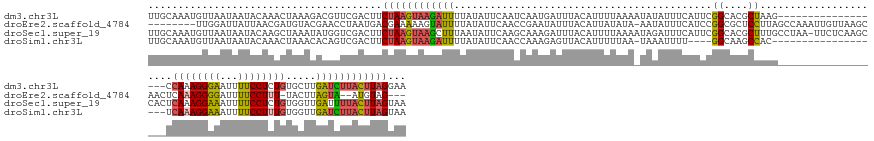
>dm3.chr3L 18316612 145 + 24543557 UUGCAAAUGUUAAUAAUACAAACUAAAGACGUUCGACUUCUAAGUAAGAUUUUAUAUUCAAUCAAUGAUUUACAUUUUAAAAUAUAUUUCAUUCGGCACGCUAAG------------------CCAAAGGGAAUUUUCCUCUGUGCUUGAUCUUACUUAGGAA ...(.((((((................)))))).)..((((((((((((((..........(((((((..((.(((....))).))..))))).))......(((------------------(((.(((((...))))).)).)))))))))))))))))). ( -27.99, z-score = -1.91, R) >droEre2.scaffold_4784 10188028 148 - 25762168 --------UUGGAUUAUUAACGAUGUACGAACCUAAUGACGAAAAAGUAUUUUAUAUUCAACCGAAUAUUUACAUUAUAUA-AAUAUUUCAUCCGGCGCUCCUUAGCCAAAUUGUUAAGCAACUCAAAGGGGAUUUUCCUUU-UACUUAGUA--AUGUAG--- --------.((.((((((((................((((((....(((....((((((....)))))).)))........-............(((........)))...))))))........(((((((....))))))-)..))))))--)).)).--- ( -23.30, z-score = 0.08, R) >droSec1.super_19 311973 162 + 1257711 UUGCAAAUGUUAAUAAUACAAGCUAAAUAUGGUCGACUUCUAAGUAAGCUUUAAUAUUCAAGCAAAGAUUUACAUUUUAAAAUAGAUUUCAUUCGGCACGCUUUGCCUAA-UUCUCAAGCCACUCAAAGGAAAUUUUCCUCUGUGGUUGAUUUUACUUAGUAA .....................(((((.((..(((((..((((.....((((........)))).(((((....)))))....))))..))....((((.....))))...-......((((((....((((.....))))..)))))))))..)).))))).. ( -28.80, z-score = -0.49, R) >droSim1.chr3L 17648952 139 + 22553184 UUGCAAAUGUUAAUAAUACAAACUAAACACAGUCGACUUCUAAGUAAGAUUUUAUAUUCAACCAAAGAGUUACAUUUUUAA-UAAAUUUU----GGCAAGCCAC-------------------UCAAAGGAAAUUUUCCUUUGUGGUUGAUCUUACUUAGUAA .......................................((((((((((((..........((((((.((((......)))-)...))))----))..((((((-------------------..((((((.....))))))))))))))))))))))))... ( -28.80, z-score = -2.59, R) >consensus UUGCAAAUGUUAAUAAUACAAACUAAACACAGUCGACUUCUAAGUAAGAUUUUAUAUUCAACCAAAGAUUUACAUUUUAAA_UAUAUUUCAUUCGGCACGCCAUG__________________UCAAAGGAAAUUUUCCUCUGUGCUUGAUCUUACUUAGUAA .......................................((((((((((((...........................................((....))......................((((((((...)))))))).....))))))))))))... (-13.46 = -13.78 + 0.31)
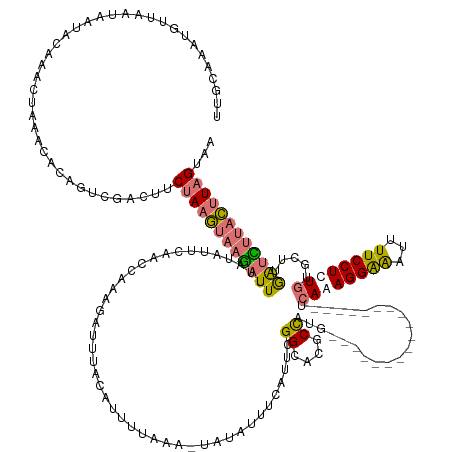
| Location | 18,316,612 – 18,316,757 |
|---|---|
| Length | 145 |
| Sequences | 4 |
| Columns | 163 |
| Reading direction | reverse |
| Mean pairwise identity | 64.99 |
| Shannon entropy | 0.55937 |
| G+C content | 0.29915 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -11.00 |
| Energy contribution | -11.62 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18316612 145 - 24543557 UUCCUAAGUAAGAUCAAGCACAGAGGAAAAUUCCCUUUGG------------------CUUAGCGUGCCGAAUGAAAUAUAUUUUAAAAUGUAAAUCAUUGAUUGAAUAUAAAAUCUUACUUAGAAGUCGAACGUCUUUAGUUUGUAUUAUUAACAUUUGCAA ...(((((((((((.((((.((((((.......)))))))------------------)))................(((((((...((((.....))))....)))))))..))))))))))).((((((((.......))))).))).............. ( -33.00, z-score = -2.92, R) >droEre2.scaffold_4784 10188028 148 + 25762168 ---CUACAU--UACUAAGUA-AAAGGAAAAUCCCCUUUGAGUUGCUUAACAAUUUGGCUAAGGAGCGCCGGAUGAAAUAUU-UAUAUAAUGUAAAUAUUCGGUUGAAUAUAAAAUACUUUUUCGUCAUUAGGUUCGUACAUCGUUAAUAAUCCAA-------- ---......--.........-...(((...(((...(..(((((.....)))))..)....)))(.(((.(((((((((((-.....)))))..((((((....))))))..........))))))....))).)...............)))..-------- ( -22.70, z-score = 0.92, R) >droSec1.super_19 311973 162 - 1257711 UUACUAAGUAAAAUCAACCACAGAGGAAAAUUUCCUUUGAGUGGCUUGAGAA-UUAGGCAAAGCGUGCCGAAUGAAAUCUAUUUUAAAAUGUAAAUCUUUGCUUGAAUAUUAAAGCUUACUUAGAAGUCGACCAUAUUUAGCUUGUAUUAUUAACAUUUGCAA ...((((((((..((((((((((((((.....))))))..)))).))))...-((((((((((..(((....(((((....)))))....)))...))))))))))..........))))))))................((.(((.......)))...)).. ( -35.90, z-score = -1.88, R) >droSim1.chr3L 17648952 139 - 22553184 UUACUAAGUAAGAUCAACCACAAAGGAAAAUUUCCUUUGA-------------------GUGGCUUGCC----AAAAUUUA-UUAAAAAUGUAACUCUUUGGUUGAAUAUAAAAUCUUACUUAGAAGUCGACUGUGUUUAGUUUGUAUUAUUAACAUUUGCAA ...((((((((((((((((((((((((.....))))))).-------------------.(((....))----).......-.................)))))))........))))))))))..(.(((.(((...((((....))))...))).)))).. ( -34.20, z-score = -3.59, R) >consensus UUACUAAGUAAGAUCAACCACAAAGGAAAAUUCCCUUUGA__________________CAAAGCGUGCCGAAUGAAAUAUA_UUUAAAAUGUAAAUCUUUGGUUGAAUAUAAAAUCUUACUUAGAAGUCGACCGUGUUUAGCUUGUAUUAUUAACAUUUGCAA ...((((((((.........((((((.......)))))).............................................................((((........))))))))))))....................................... (-11.00 = -11.62 + 0.62)

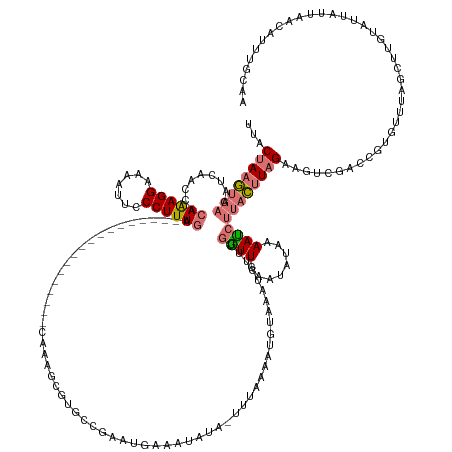
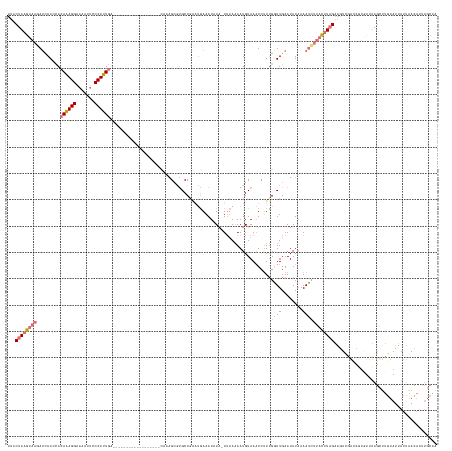
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:35 2011