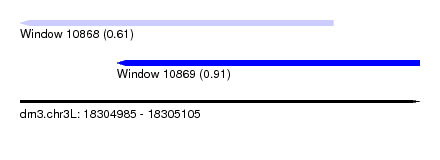
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,304,985 – 18,305,105 |
| Length | 120 |
| Max. P | 0.914163 |

| Location | 18,304,985 – 18,305,079 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.44 |
| Shannon entropy | 0.37080 |
| G+C content | 0.38605 |
| Mean single sequence MFE | -18.64 |
| Consensus MFE | -10.22 |
| Energy contribution | -9.90 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.608915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
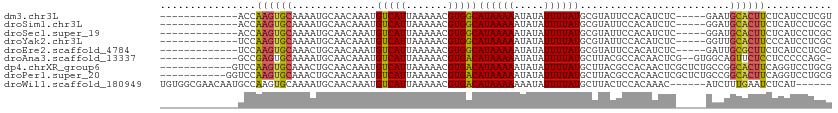
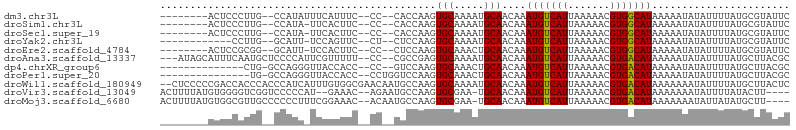
>dm3.chr3L 18304985 94 - 24543557 -------------ACCAAGUGCAAAAUGCAACAAAUGUCAUUAAAAACGUGGCAUAAAAAUAUAUUUUAUGCGUAUUCCACAUCUC-----GAAUGCACUUCUCAUCCUCGU -------------...(((((((.((((((.....)).))))......((((.(((...(((.....)))...))).)))).....-----...)))))))........... ( -17.30, z-score = -2.44, R) >droSim1.chr3L 17637396 94 - 22553184 -------------ACCAAGUGCAAAAUGCAACAAAUGUCAUUAAAAACGUGGCAUAAAAAUAUAUUUUAUGCGUAUUCCACAUCUC-----GGAUGCACUUCUCAUCCUCGC -------------...(((((((..((((.....((((........)))).((((((((.....))))))))))))(((.......-----))))))))))........... ( -19.30, z-score = -2.53, R) >droSec1.super_19 300516 94 - 1257711 -------------ACCAAGUGCAAAAUGCAACAAAUGUCAUUAAAAACGUGGCAUAAAAAUAUAUUUUAUGCGUAUUCCACAUCUC-----GGAUGCACUUCUCAUCCUCGC -------------...(((((((..((((.....((((........)))).((((((((.....))))))))))))(((.......-----))))))))))........... ( -19.30, z-score = -2.53, R) >droYak2.chr3L 8459659 94 + 24197627 -------------UCCAAGUGCAAAAUGCAACAAAUGUCAUUAAAAACGUGGCAUAAAAAUAUAUUUUAUGCGUAUUCCACAUCUC-----GGUUGCACUUCCCAUCCUCGC -------------...(((((((((((((.....((((........)))).((((((((.....)))))))))))))((.......-----))))))))))........... ( -19.00, z-score = -2.55, R) >droEre2.scaffold_4784 10176671 94 + 25762168 -------------UCCAAGUGCAAACUGCAACAAAUGUCAUUAAAAACGUGGCAUAAAAAUAUAUUUUAUGCGUAUUCCACAUCUC-----GAUUGCGCUUCUCAUCCUCGC -------------...((((((((..........((((........)))).((((((((.....))))))))..............-----..))))))))........... ( -16.30, z-score = -1.54, R) >droAna3.scaffold_13337 4207391 96 + 23293914 -------------GCCGAGUGCAAAAUGCAACAAAUGUCAUUAAAAACGUGACAUAAAAAUAUAUUUUAUGCUUACGCCACAACUCG--GUGGCAGUUCUCCUCCCCCAGC- -------------(((((((((.....))..................((((((((((((.....))))))).))))).....)))))--))((.((.....)).)).....- ( -20.90, z-score = -2.02, R) >dp4.chrXR_group6 11183522 100 - 13314419 ------------GUCCAAGUGCAAACUGCAACAAAUGUCAUUAAAAACGUGACAUAAAAAUAUAUUUUAUGCUUACGCCACAACUCGCUCUGCCGGCACUUCAGGUCCUGCG ------------(.((((((((.....(((..................(((((((((((.....))))))).))))((........))..)))..))))))..)).)..... ( -17.90, z-score = -0.48, R) >droPer1.super_20 1084783 101 - 1872136 -----------GGUCCAAGUGCAAACUGCAACAAAUGUCAUUAAAAACGUGACAUAAAAAUAUAUUUUAUGCUUACGCCACAACUCGCUCUGCCGGCACUUCAGGUCCUGCG -----------((.((((((((.....(((..................(((((((((((.....))))))).))))((........))..)))..))))))..)).)).... ( -21.60, z-score = -1.35, R) >droWil1.scaffold_180949 5744269 100 + 6375548 UGUGGCGAACAAUGCCAAGUGCAAAAUGCAACAAAUGUCAUUAAAAACGUGACAUAAAAAAAUAUUUUAUGCUUACUCCACAAAC------AUCUUUGAAUCUCAU------ (((((.....((((.((..(((.....))).....)).))))......(((((((((((.....))))))).)))).)))))...------...............------ ( -16.20, z-score = -1.03, R) >consensus _____________UCCAAGUGCAAAAUGCAACAAAUGUCAUUAAAAACGUGGCAUAAAAAUAUAUUUUAUGCGUAUUCCACAUCUC_____GGCUGCACUUCUCAUCCUCGC ................((((((..............(((((.......)))))((((((.....)))))).........................))))))........... (-10.22 = -9.90 + -0.32)
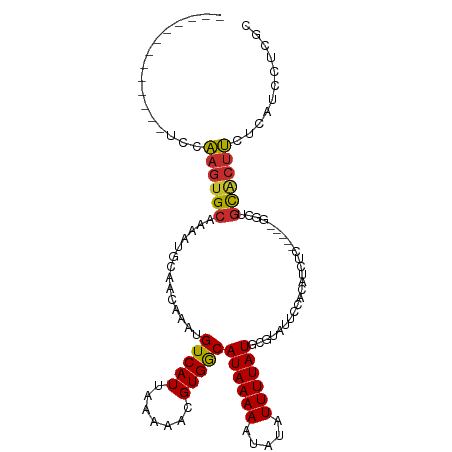
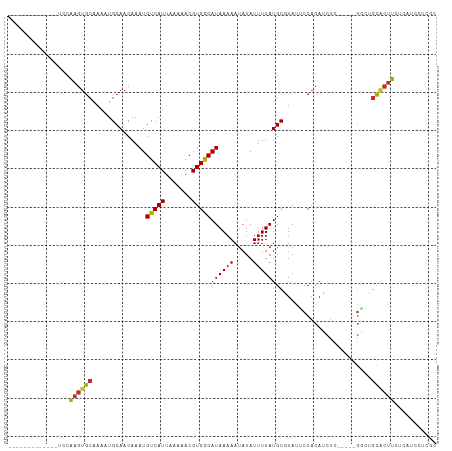
| Location | 18,305,014 – 18,305,105 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 72.28 |
| Shannon entropy | 0.58938 |
| G+C content | 0.37151 |
| Mean single sequence MFE | -17.13 |
| Consensus MFE | -8.05 |
| Energy contribution | -7.80 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18305014 91 - 24543557 --------ACUCCCUUG--CCAUAUUUCAUUUC--CC--CACCAAGUGCAAAAUGCAACAAAUGUCAUUAAAAACGUGGCAUAAAAAUAUAUUUUAUGCGUAUUC --------.......((--(((((.........--..--.......(((.....)))....(((((((.......)))))))............)))).)))... ( -11.10, z-score = -0.51, R) >droSim1.chr3L 17637425 90 - 22553184 --------ACUCCCUUG--CCAUA-UUCACUUC--CC--CACCAAGUGCAAAAUGCAACAAAUGUCAUUAAAAACGUGGCAUAAAAAUAUAUUUUAUGCGUAUUC --------......(((--(....-..(((((.--..--....)))))......))))...((((........)))).((((((((.....))))))))...... ( -12.40, z-score = -1.22, R) >droSec1.super_19 300545 90 - 1257711 --------ACUCCCUUG--CCAUA-UUCACUUC--CC--CACCAAGUGCAAAAUGCAACAAAUGUCAUUAAAAACGUGGCAUAAAAAUAUAUUUUAUGCGUAUUC --------......(((--(....-..(((((.--..--....)))))......))))...((((........)))).((((((((.....))))))))...... ( -12.40, z-score = -1.22, R) >droYak2.chr3L 8459688 86 + 24197627 ------------CCUUG--GCAUU-UCCAGUUC--CU--CUCCAAGUGCAAAAUGCAACAAAUGUCAUUAAAAACGUGGCAUAAAAAUAUAUUUUAUGCGUAUUC ------------.....--(((((-(..((...--..--))..))))))..(((((.....((((........)))).((((((((.....))))))))))))). ( -15.20, z-score = -1.61, R) >droEre2.scaffold_4784 10176700 90 + 25762168 --------ACUCCGCGG--GCAUU-UCCACUUC--CC--CUCCAAGUGCAAACUGCAACAAAUGUCAUUAAAAACGUGGCAUAAAAAUAUAUUUUAUGCGUAUUC --------....((((.--(((((-(.(((((.--..--....))))).))).))).((....)).........))))((((((((.....))))))))...... ( -17.10, z-score = -1.51, R) >droAna3.scaffold_13337 4207422 98 + 23293914 ---AUAGCAUUUCAAUGCUCCCCAUUCGUUUUU--CC--CGCCGAGUGCAAAAUGCAACAAAUGUCAUUAAAAACGUGACAUAAAAAUAUAUUUUAUGCUUACGC ---...((((((...(((....((((((.....--..--...))))))......)))..)))))).........((((((((((((.....))))))).))))). ( -17.90, z-score = -1.98, R) >dp4.chrXR_group6 11183556 86 - 13314419 --------------CUG-GCCAGGGUUACCACC--CC--GUCCAAGUGCAAACUGCAACAAAUGUCAUUAAAAACGUGACAUAAAAAUAUAUUUUAUGCUUACGC --------------.((-(.(.((((....)))--).--).)))..(((.....))).................((((((((((((.....))))))).))))). ( -18.60, z-score = -1.42, R) >droPer1.super_20 1084817 87 - 1872136 ---------------UG-GCCAGGGUUACCACC--CCUGGUCCAAGUGCAAACUGCAACAAAUGUCAUUAAAAACGUGACAUAAAAAUAUAUUUUAUGCUUACGC ---------------((-(((((((.......)--))))).)))..(((.....))).................((((((((((((.....))))))).))))). ( -24.80, z-score = -2.92, R) >droWil1.scaffold_180949 5744291 103 + 6375548 --CUCCCCCGACCACCCACCCAUCAUUUGUGGCGAACAAUGCCAAGUGCAAAAUGCAACAAAUGUCAUUAAAAACGUGACAUAAAAAAAUAUUUUAUGCUUACUC --.....................(((((((((((.....))))...(((.....))))))))))...........(((((((((((.....))))))).)))).. ( -18.40, z-score = -2.26, R) >droVir3.scaffold_13049 21494244 96 - 25233164 ACUUUUAUGUGGGGUCGGUCCCCCAU--GAAAC--AGAAUGCCAAGUGCGAA-UGCAACAAAUGUCAUUAAAAACGUGACAUAAAAAAAUAUUUUAUACUU---- ..(((((((.((((.....)))))))--)))).--((((((.....(((...-.)))....(((((((.......))))))).......))))))......---- ( -21.30, z-score = -1.57, R) >droMoj3.scaffold_6680 15928169 98 - 24764193 ACUUUUAUGUGGCGUUGCCCCCCUUUCGGAAAC--ACAAUGCCAAGUGCGAA-UGCAACAAAUGUCAUUAAAAACGUGACAUAAAAAAAUAUUAUAUGCUU---- ..((((((((.(((((...........(....)--..((((.((..(((...-.))).....)).))))...))))).))))))))...............---- ( -19.20, z-score = -1.03, R) >consensus ________ACUCCCUUG__CCAUA_UUCACUUC__CC__CGCCAAGUGCAAAAUGCAACAAAUGUCAUUAAAAACGUGACAUAAAAAUAUAUUUUAUGCUUAUUC ..............................................(((.....)))....(((((((.......)))))))....................... ( -8.05 = -7.80 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:33 2011