| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,280,497 – 18,280,592 |
| Length | 95 |
| Max. P | 0.998603 |

| Location | 18,280,497 – 18,280,592 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.35 |
| Shannon entropy | 0.54009 |
| G+C content | 0.37580 |
| Mean single sequence MFE | -25.29 |
| Consensus MFE | -18.01 |
| Energy contribution | -17.71 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.998603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
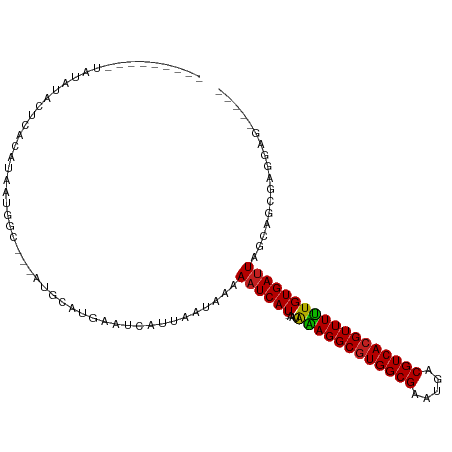
>dm3.chr3L 18280497 95 - 24543557 ---------UAUAUACUCGCAUAAUGGC---AUGCAUGAAUCAUUAAUAAAAAUCAU-AAUAAAGGCGUGGCGAAUGACGUCACGUUUUUGUGAUUAGCCACGAGGAG----- ---------......((((.....((((---....((((..............))))-.((((((((((((((.....)))))))))))))).....))))))))...----- ( -26.74, z-score = -2.93, R) >droGri2.scaffold_14385 87777 103 - 112342 AAUUCUCGCUAUAUGCAUAUGCAAUAGU--GGCCAG-GCAUCAUUAAUAAAAAUCAU-AAUAAAGGCGUGGCGAAUGACGUCACGUUUUUGUGAUUAGCACAGACAG------ .....(((((((.(((....))))))))--))....-..............((((((-((...((((((((((.....))))))))))))))))))...........------ ( -27.10, z-score = -2.08, R) >droMoj3.scaffold_6680 15901559 105 - 24764193 ----CACGCUUUAUGCAUAUGUAAUAGC--AAUGCG-GCAUCAUUAAUAAAAAUCAU-AAUAAAGGCGUGGCGAAUGACGUCACGUUUUUGUGAUUAGCACGGAGACGGAGAG ----..(((((..(((..((((..((..--..))..-))))..........((((((-((...((((((((((.....)))))))))))))))))).)))..))).))..... ( -30.60, z-score = -2.99, R) >droVir3.scaffold_13049 21471429 103 - 25233164 AAUAUUCGCUUUAUGCAUAUGAAAUAGU--AACGCG-GCAUCAUUAAUAAAAAUCAU-AAUAAAGGCGUGGCGAAUGACGUCACGUUUUUGUGAUUAGCAGAGACAG------ ....((((((((((....((((....(.--....).-...))))..)))))((((((-((...((((((((((.....)))))))))))))))))).)).)))....------ ( -23.00, z-score = -1.11, R) >droWil1.scaffold_180949 5712058 99 + 6375548 ----AAUAAGCUAAAGUAGCAGCAUAUUUUGAAUUAAAGAUCAUUAAUAAAAAUCAUAAAUAAAGGCAUGGCGAAUGACGUCACGUUUUUGUGAUUAGCUGAG---------- ----...............((((.((((.(((........)))..))))..(((((((((....(((.(((((.....))))).)))))))))))).))))..---------- ( -19.10, z-score = -1.26, R) >droPer1.super_20 1059813 91 - 1872136 ---------------UGUAUGGUUUGGU-AUGCAUA-GGAUCAUUAAUAAAAAUCAU-AAUGGAGGCGUGGCGAAUGACGUCACGUUUCUGUGAUUAGCAGAGACCGAG---- ---------------........(((((-.(((.((-(.....))).....((((((-...((((((((((((.....)))))))))))))))))).)))...))))).---- ( -28.30, z-score = -3.34, R) >dp4.chrXR_group6 11159290 91 - 13314419 ---------------UGUAUGGUUUGGU-AUGCAUA-GGAUCAUUAAUAAAAAUCAU-AAUGGAGGCGUGGCGAAUGACGUCACGUUUCUGUGAUUAGCAGAGACCGAG---- ---------------........(((((-.(((.((-(.....))).....((((((-...((((((((((((.....)))))))))))))))))).)))...))))).---- ( -28.30, z-score = -3.34, R) >droAna3.scaffold_13337 4185404 91 + 23293914 ---------AAUAUAUUACCAUAAUGCC-CUACACAUGAAUCAUUAAUAAAAAUCAU-AAUAGAGGCGUGGCGAAUGACGUCACGUUUUUGUGAUUAGCUUC----------- ---------...................-......................((((((-(...(((((((((((.....))))))))))))))))))......----------- ( -18.70, z-score = -1.64, R) >droEre2.scaffold_4784 10153222 95 + 25762168 ---------UAUAUAGUCACAUAAUGGC---AUGCAUGAAUCAUUAAUAAAAAUCAU-AAUAAAGGCGUGGCGAAUGACGUCACGUUUUUGUGAUUAGCCUCGUGGAG----- ---------....(((((((.(((((.(---(....))...)))))...........-...((((((((((((.....))))))))))))))))))).((....))..----- ( -24.30, z-score = -2.02, R) >droYak2.chr3L 8435003 95 + 24197627 ---------UAUAUACUCAUAUAAUGGC---AUGCAUGAAUCAUUAAUAAAAAUCAU-AAUAAAGGCGUGGCGAAUGACGUCACGUUUUUGUGACUAGCCUCGUGGAG----- ---------......(((.(((...(((---....((((..............))))-.((((((((((((((.....)))))))))))))).....)))..))))))----- ( -23.84, z-score = -2.05, R) >droSec1.super_19 274654 95 - 1257711 ---------AAUAUACUCGCAUAAUGGC---AUGCAUGAAUCAUUAAUAAAAAUCAU-AAUAAAGGCGUGGCGAAUGACGUCACGUUUUUGUGAUUAGCCACGAGGAG----- ---------......((((.....((((---....((((..............))))-.((((((((((((((.....)))))))))))))).....))))))))...----- ( -26.74, z-score = -2.95, R) >droSim1.chr3L 17609754 95 - 22553184 ---------AAUAUACUCGCAUAAUGGC---AUGCAUGAAUCAUUAAUAAAAAUCAU-AAUAAAGGCGUGGCGAAUGACGUCACGUUUUUGUGAUUAGCCACGAGGAG----- ---------......((((.....((((---....((((..............))))-.((((((((((((((.....)))))))))))))).....))))))))...----- ( -26.74, z-score = -2.95, R) >consensus _________UAUAUACUCACAUAAUGGC___AUGCAUGAAUCAUUAAUAAAAAUCAU_AAUAAAGGCGUGGCGAAUGACGUCACGUUUUUGUGAUUAGCAGCGAGGAG_____ ...................................................((((((....((((((((((((.....))))))))))))))))))................. (-18.01 = -17.71 + -0.30)

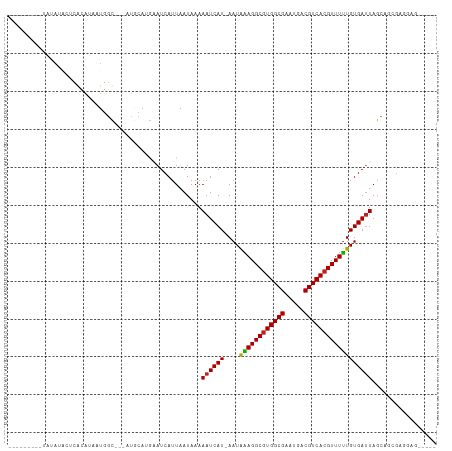
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:31 2011