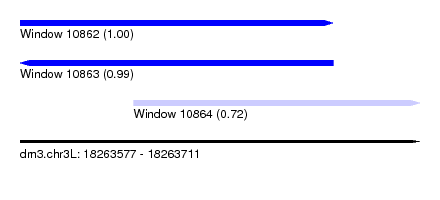
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,263,577 – 18,263,711 |
| Length | 134 |
| Max. P | 0.995798 |

| Location | 18,263,577 – 18,263,682 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 67.57 |
| Shannon entropy | 0.63779 |
| G+C content | 0.33938 |
| Mean single sequence MFE | -25.41 |
| Consensus MFE | -7.84 |
| Energy contribution | -9.37 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.995798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
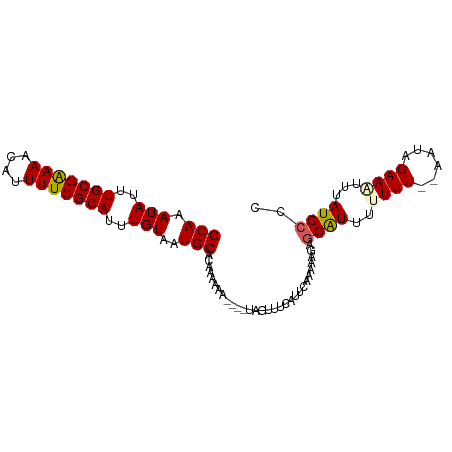
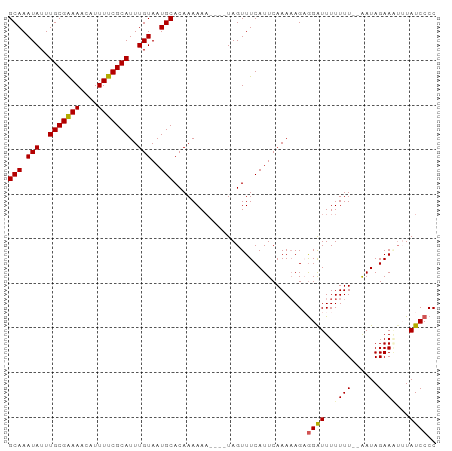
>dm3.chr3L 18263577 105 + 24543557 GUUGAACGUGGAGGGGGUUAAUUCCACUCCACAACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAAAAAAAAAUAGUUUCAUUCAAAAAGA .(((((.((((((.(((.....))).))))))...((((((.(((..(((((((....)))))))..)))..)))))).................)))))..... ( -33.50, z-score = -5.17, R) >droSim1.chr3L 17592886 100 + 22553184 GUUGUGUGUGGAGGGGGUUAAUUCCACUCCACAACUGUGCAAAUAUUUGCGGAAACAUUUUCGCAUUUGUAAUGCACAAAAA-----UAGUUUCAUUCAAAAAGU ((((((.((((((........))))))..))))))((((((.(((..(((((((....)))))))..)))..))))))....-----.................. ( -31.40, z-score = -3.78, R) >droSec1.super_19 257663 101 + 1257711 GUUGUGUGUGGAGAGGGUUAAUUCCACUCCACAACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAAAAA----UAGUUUCAUUCAAAAAGU ......(((((((.(((.....))).)))))))..((((((.(((..(((((((....)))))))..)))..)))))).....----.................. ( -31.60, z-score = -4.35, R) >droYak2.chr3L 8417444 96 - 24197627 GUUUUGCGUGGAGGGAGUUAAUCCCAC-----AACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAAAAA----UAGUUUCAUUCAAAAAGA .(((((.(((((((((.....))))..-----...((((((.(((..(((((((....)))))))..)))..)))))).....----....))))).)))))... ( -29.80, z-score = -4.28, R) >droEre2.scaffold_4784 10136705 96 - 25762168 GUUGAGCGUGGAGAGAGUUAAUACCAC-----AACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAAAAA----UAGUUUCAUUCAAAUAGA .(((((.((((............))))-----...((((((.(((..(((((((....)))))))..)))..)))))).....----........)))))..... ( -23.20, z-score = -2.36, R) >dp4.chrXR_group6 11136119 81 + 13314419 GCUAGAGCGAGAGAGAGUGUGUUGGAUCAAACAACUGUGCAAAUAAUUGCAGAAAAAUAACCA-AUAUAUUUUGAAGAGAAU----------------------- ((....)).....((((((((((((.....((....))((((....))))..........)))-))))))))).........----------------------- ( -15.50, z-score = -2.27, R) >triCas2.ChLG3 26746469 92 + 32080666 GUAAGACGGGAGGAGUUUUAGCAGAACU---UCAGUCU-CCAAUAUUUGUAAAAAAAUUU----GUUUUUAACGAGAAACUUA-----AAAUACAAGAGAAAAAA ...((((....(((((((.....)))))---)).))))-......((((((.........----((((((....))))))...-----...))))))........ ( -12.86, z-score = 0.07, R) >consensus GUUGUGCGUGGAGAGAGUUAAUUCCACU__ACAACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAAAAA____UAGUUUCAUUCAAAAAGA ...................................((((((.(((..(((((((....)))))))..)))..))))))........................... ( -7.84 = -9.37 + 1.53)


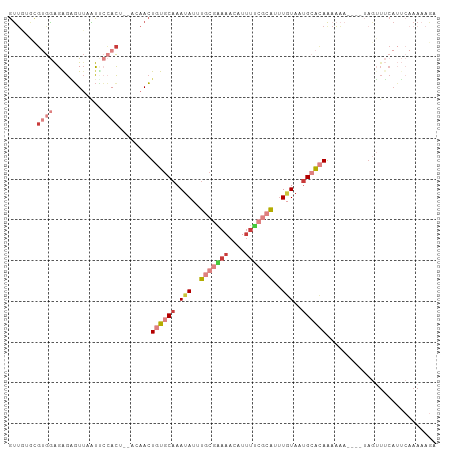
| Location | 18,263,577 – 18,263,682 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 67.57 |
| Shannon entropy | 0.63779 |
| G+C content | 0.33938 |
| Mean single sequence MFE | -21.03 |
| Consensus MFE | -8.39 |
| Energy contribution | -11.06 |
| Covariance contribution | 2.68 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.992610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18263577 105 - 24543557 UCUUUUUGAAUGAAACUAUUUUUUUUUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUUGUGGAGUGGAAUUAACCCCCUCCACGUUCAAC .....((((((...............(((((((.......(((((((....)))))))......)))))))..((((((.((........)))))))))))))). ( -31.92, z-score = -4.91, R) >droSim1.chr3L 17592886 100 - 22553184 ACUUUUUGAAUGAAACUA-----UUUUUGUGCAUUACAAAUGCGAAAAUGUUUCCGCAAAUAUUUGCACAGUUGUGGAGUGGAAUUAACCCCCUCCACACACAAC ...........(((((.(-----(((((((...........))))))))))))).((((....))))...(((((((((.((........))))))))).))... ( -25.80, z-score = -3.14, R) >droSec1.super_19 257663 101 - 1257711 ACUUUUUGAAUGAAACUA----UUUUUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUUGUGGAGUGGAAUUAACCCUCUCCACACACAAC ..................----.....((((((.......(((((((....)))))))......))))))(((((((((.((.......)).))))))).))... ( -29.82, z-score = -4.42, R) >droYak2.chr3L 8417444 96 + 24197627 UCUUUUUGAAUGAAACUA----UUUUUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUU-----GUGGGAUUAACUCCCUCCACGCAAAAC ...(((((..((......----....(((((((.......(((((((....)))))))......)))))))..-----..((((.....))))..))..))))). ( -23.92, z-score = -3.01, R) >droEre2.scaffold_4784 10136705 96 + 25762168 UCUAUUUGAAUGAAACUA----UUUUUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUU-----GUGGUAUUAACUCUCUCCACGCUCAAC .....((((.........----....(((((((.......(((((((....)))))))......)))))))..-----((((............))))..)))). ( -21.12, z-score = -2.50, R) >dp4.chrXR_group6 11136119 81 - 13314419 -----------------------AUUCUCUUCAAAAUAUAU-UGGUUAUUUUUCUGCAAUUAUUUGCACAGUUGUUUGAUCCAACACACUCUCUCUCGCUCUAGC -----------------------.................(-((((((..(..((((((....))))..))..)..))).))))..................... ( -7.10, z-score = 0.07, R) >triCas2.ChLG3 26746469 92 - 32080666 UUUUUUCUCUUGUAUUU-----UAAGUUUCUCGUUAAAAAC----AAAUUUUUUUACAAAUAUUGG-AGACUGA---AGUUCUGCUAAAACUCCUCCCGUCUUAC (((..(((((.((((((-----...(.....)((.(((((.----....))))).)))))))).))-)))..))---)........................... ( -7.50, z-score = 1.00, R) >consensus UCUUUUUGAAUGAAACUA____UUUUUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUUGU__AGUGGAAUUAACCCUCUCCACGCACAAC ..........................(((((((.......(((((((....)))))))......))))))).......(((((..........)))))....... ( -8.39 = -11.06 + 2.68)

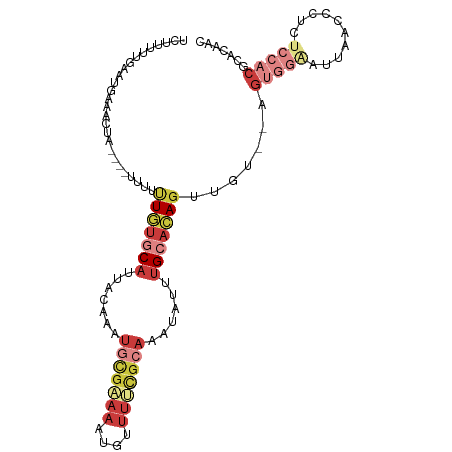
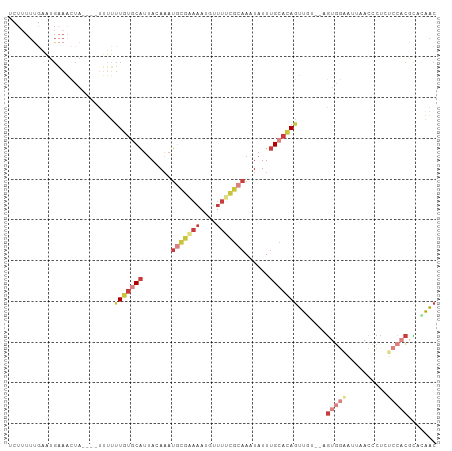
| Location | 18,263,615 – 18,263,711 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 91.59 |
| Shannon entropy | 0.14736 |
| G+C content | 0.27863 |
| Mean single sequence MFE | -16.42 |
| Consensus MFE | -13.28 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.722801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18263615 96 + 24543557 GCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAAAAAAAAAUAGUUUCAUUCAAAAAGAGGAUUUUUUUGUAAUAGAAAUGUAUCCCC (((.(((..(((((((....)))))))..)))..)))..............((((((((((((((((...)))))))).))).)))))........ ( -18.30, z-score = -2.30, R) >droSim1.chr3L 17592924 89 + 22553184 GCAAAUAUUUGCGGAAACAUUUUCGCAUUUGUAAUGCACAAAAA-----UAGUUUCAUUCAAAAAGUGGAUUUUUUU--AAUAGAAGUUUAUCCCC (((.(((..(((((((....)))))))..)))..))).......-----................((((((((((..--...)))))))))).... ( -15.50, z-score = -1.37, R) >droSec1.super_19 257701 90 + 1257711 GCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAAAAA----UAGUUUCAUUCAAAAAGUAGAUUUUUUU--AAUAGAAAUAUAUCCCC (((.(((..(((((((....)))))))..)))..)))........----..((((((((.((((((.....))))))--))).)))))........ ( -15.50, z-score = -2.71, R) >droYak2.chr3L 8417477 90 - 24197627 GCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAAAAA----UAGUUUCAUUCAAAAAGAGGGUUUUUUU--GAUAGAAUUUUAUCCCC (((.(((..(((((((....)))))))..)))..)))........----....(((..(((((((((...)))))))--))..))).......... ( -17.40, z-score = -1.88, R) >droEre2.scaffold_4784 10136738 91 - 25762168 GCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAAAAA----UAGUUUCAUUCAAAUAGAGGGUUUUUUUU-AAUAGAAUGUUAUCCCC (((.(((..(((((((....)))))))..)))..)))........----..((..(((((...((((((....)))))-)...)))))..)).... ( -15.40, z-score = -1.32, R) >consensus GCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAAAAA____UAGUUUCAUUCAAAAAGAGGAUUUUUUU__AAUAGAAAUUUAUCCCC (((.(((..(((((((....)))))))..)))..)))..............................((((..((((......))))...)))).. (-13.28 = -13.40 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:29 2011