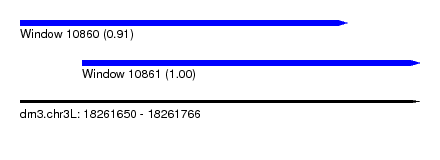
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,261,650 – 18,261,766 |
| Length | 116 |
| Max. P | 0.998307 |

| Location | 18,261,650 – 18,261,745 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.83 |
| Shannon entropy | 0.36582 |
| G+C content | 0.51319 |
| Mean single sequence MFE | -37.36 |
| Consensus MFE | -17.47 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
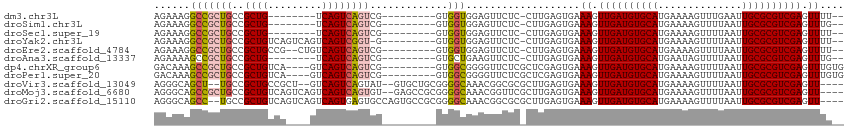
>dm3.chr3L 18261650 95 + 24543557 AGAAAGGCCGCUGCCGCUG--------UCAGUCAGUCG---------GUGGUGGAGUUCUC-CUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUGAAUUGCGCGUCGAGUUUU-- ((((...((((..((((((--------.....))).))---------)..))))..)))).-.......((((.((((((((((..............)))))))))).))))-- ( -35.94, z-score = -3.79, R) >droSim1.chr3L 17590912 95 + 22553184 AGAAAGGCCGCUGCCGCUG--------UCAGUCAGUCG---------GUGGUGGAGUUCUC-CUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUUG-- ((((...((((..((((((--------.....))).))---------)..))))..)))).-........(((.((((((((((..............)))))))))).))).-- ( -35.54, z-score = -3.89, R) >droSec1.super_19 255729 95 + 1257711 AGAAAGGCCGCUGCCGCUG--------UCAGUCAGUCG---------GUGGUGGAGUUCUC-CUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUUU-- ((((...((((..((((((--------.....))).))---------)..))))..)))).-.......((((.((((((((((..............)))))))))).))))-- ( -35.94, z-score = -4.01, R) >droYak2.chr3L 8415538 102 - 24197627 AGAAAGGCCGCUGCCGCUGUCAGUCAGUCAGUCGGU-G---------GUGGUGGAGUUCUC-CUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUUU-- ......(((((..((((((.(.....).))).))).-.---------)))))(((....))-)......((((.((((((((((..............)))))))))).))))-- ( -38.04, z-score = -3.96, R) >droEre2.scaffold_4784 10134897 101 - 25762168 AGAAAGGCCGCUGCCGCUGCCG--CUGUCAGUCAGUCG---------GUGGUGGAGUUCUC-CUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUUU-- ...((((..(((.((((..(((--(((.....))).))---------)..)))))))...)-)))....((((.((((((((((..............)))))))))).))))-- ( -38.74, z-score = -3.52, R) >droAna3.scaffold_13337 4168103 95 - 23293914 AGAAAAGCCGCUGCCGCUG--------UCAGUCAGUCG---------GUGCUGAAGUUCUC-CUUGAGUGAAAGUUGAUGUGCAUGAAUAGUUUUAAUUGCGCGUCGAGUUUG-- ((((...(.((.(((((((--------.....))).))---------)))).)...)))).-........(((.((((((((((((((....))))..)))))))))).))).-- ( -28.00, z-score = -1.72, R) >dp4.chrXR_group6 11133693 102 + 13314419 GACAAAGCCGCUGCCGCUGUCA----GUCAGUCAGUCG---------GUGGCGGGGUUCUCGCUCGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUUGUG .(((((.((((..((((((.(.----....).))).))---------)..)))).....((((....))))...((((((((((..............)))))))))).))))). ( -38.14, z-score = -2.48, R) >droPer1.super_20 1032828 102 + 1872136 GACAAAGCCGCUGCCGCUGUCA----GUCAGUCAGUCG---------GUGGCGGGGUUCUCGCUCGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUUGUG .(((((.((((..((((((.(.----....).))).))---------)..)))).....((((....))))...((((((((((..............)))))))))).))))). ( -38.14, z-score = -2.48, R) >droVir3.scaffold_13049 21451459 105 + 25233164 AGGGCAGCU--UGCCGCUGCCGCU--GUCAGUCAGUAU--GUGCUGCGGGGCAAACGGCGCGCUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUU---- ..((((((.--....))))))(((--.(((.((((...--((((.((.(......).)))))))))).))).)))(((((((((..............)))))))))....---- ( -39.34, z-score = -0.49, R) >droMoj3.scaffold_6680 15876802 109 + 24764193 AGGGCAGCCGCUGCCGCUGUCAGUCAGUCAGUCAGUGU--GAGCCGCGGGGCAAACGGUUCGCUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUU---- ..((((((.......))))))......(((.((((.((--((((((.........)))))))))))).)))((.((((((((((..............)))))))))).))---- ( -39.54, z-score = -1.33, R) >droGri2.scaffold_15110 2412425 109 - 24565398 AGGGCAGCC--UGCCGCUGUCAGUCAGUCAGUGAGUGCCAGUGCCGCGGGGCAAACGGCGCGCUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUU---- ..((((((.--....))))))..(((.((((.(.(((((..((((....))))...))))).))))).)))((.((((((((((..............)))))))))).))---- ( -43.64, z-score = -1.86, R) >consensus AGAAAAGCCGCUGCCGCUGUCA____GUCAGUCAGUCG_________GUGGUGGAGUUCUC_CUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUUG__ ......(((((((..((((.(.....).)))))))).............)))...................((.((((((((((..............)))))))))).)).... (-17.47 = -17.70 + 0.23)
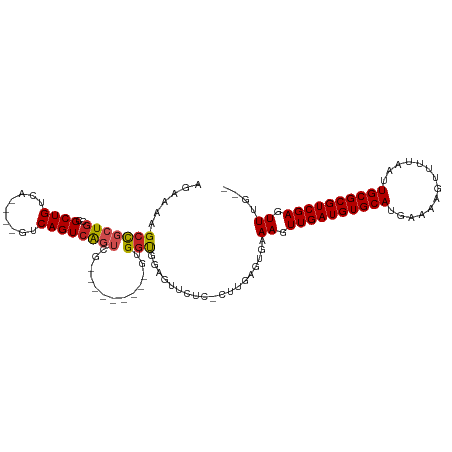
| Location | 18,261,668 – 18,261,766 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.83 |
| Shannon entropy | 0.38521 |
| G+C content | 0.49909 |
| Mean single sequence MFE | -38.31 |
| Consensus MFE | -21.05 |
| Energy contribution | -20.24 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.57 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18261668 98 + 24543557 --GUCAGUCAGUCGGUGGUGG-------------AGUUCUC-CUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUGAAUUGCGCGUCGAGUUUU--GCUCGGCAUCUCUGGGUGGCA --((((.((((..((((..((-------------(....))-)(((((..(((.((((((((((..............)))))))))).))).--.))))))))).)))).)))). ( -40.74, z-score = -4.82, R) >droSim1.chr3L 17590930 98 + 22553184 --GUCAGUCAGUCGGUGGUGG-------------AGUUCUC-CUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUUG--GCUCGGCAUCUCUGGGUGGCA --((((.((((..((((..((-------------(....))-)((((((.(((.((((((((((..............)))))))))).))).--)))))))))).)))).)))). ( -39.74, z-score = -4.66, R) >droSec1.super_19 255747 98 + 1257711 --GUCAGUCAGUCGGUGGUGG-------------AGUUCUC-CUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUUU--GCUCGGCAUCUCUGGGUGGCA --((((.((((..((((..((-------------(....))-)(((((..(((.((((((((((..............)))))))))).))).--.))))))))).)))).)))). ( -40.74, z-score = -5.11, R) >droYak2.chr3L 8415560 101 - 24197627 --GUCAGUCAGUCGGUGGUGGU----------GGAGUUCUC-CUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUUU--GCUCGGCAUCUCUGGGUGGUA --.....(((.((((.((((..----------(((....))-)(((((..(((.((((((((((..............)))))))))).))).--.))))))))).)))).))).. ( -38.14, z-score = -4.70, R) >droEre2.scaffold_4784 10134919 100 - 25762168 CUGUCAGUCAGUCGGUGGUGG-------------AGUUCUC-CUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUUU--GCUCGGCAUCUCUGGGUGGCA ..((((.((((..((((..((-------------(....))-)(((((..(((.((((((((((..............)))))))))).))).--.))))))))).)))).)))). ( -40.74, z-score = -4.85, R) >droAna3.scaffold_13337 4168121 98 - 23293914 --GUCAGUCAGUCGGUGCUGA-------------AGUUCUC-CUUGAGUGAAAGUUGAUGUGCAUGAAUAGUUUUAAUUGCGCGUCGAGUUUG--GCCCUGCAUCUCUGGGUGGCA --((((.((((..(((((...-------------...((..-...))((.(((.((((((((((((((....))))..)))))))))).))).--))...))))).)))).)))). ( -34.80, z-score = -2.89, R) >dp4.chrXR_group6 11133715 101 + 13314419 --GUCAGUCAGUCGGUGGCGG-------------GGUUCUCGCUCGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUUGUGGCCCUGCAACACUGGGUGGCA --....((((.((((((((((-------------(((((((....)))..(((.((((((((((..............)))))))))).)))..))))))))..)))))).)))). ( -42.54, z-score = -3.71, R) >droPer1.super_20 1032850 101 + 1872136 --GUCAGUCAGUCGGUGGCGG-------------GGUUCUCGCUCGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUUGUGGCCCUGCAACACUGGGUGGCA --....((((.((((((((((-------------(((((((....)))..(((.((((((((((..............)))))))))).)))..))))))))..)))))).)))). ( -42.54, z-score = -3.71, R) >droVir3.scaffold_13049 21451479 101 + 25233164 ----CUGUCAGUCAGUAUGUG--CUGCGGGGCAAACGGCGCGCUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUGG---------AGUUUUUAGUGGCA ----..((((.(((.(..(((--(.((.(......).))))))...).)))((.((((((((((..............)))))))))).))..---------.........)))). ( -28.94, z-score = -0.06, R) >droMoj3.scaffold_6680 15876824 104 + 24764193 --GUCAGUCAGUCAGUGUGAG--CCGCGGGGCAAACGGUUCGCUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUGAA--------GAUUUUUAGUGGCA --((((.(((.((((.(((((--(((.........)))))))))))).)))((.((((((((((..............)))))))))).))...--------.........)))). ( -35.64, z-score = -2.89, R) >droGri2.scaffold_15110 2412445 104 - 24565398 --GUCAGUCAGUGAGUGCCAGUGCCGCGGGGCAAACGGCGCGCUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUG----------GAUUUUUAGUGGCA --....((((.((((((((..((((....))))...)))))....((((..((.((((((((((..............)))))))))).)).----------.))))))).)))). ( -36.84, z-score = -1.83, R) >consensus __GUCAGUCAGUCGGUGGUGG_____________AGUUCUC_CUUGAGUGAAAGUUGAUGUGCAUGAAAAGUUUUAAUUGCGCGUCGAGUUUG__GCUCGGCAUCUCUGGGUGGCA ......((((.((((.(.....................(((....)))...((.((((((((((..............)))))))))).)).............).)))).)))). (-21.05 = -20.24 + -0.81)

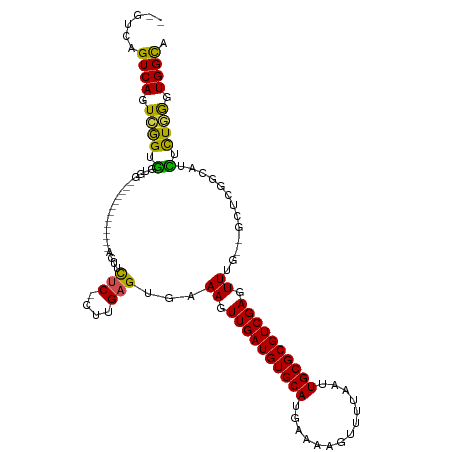
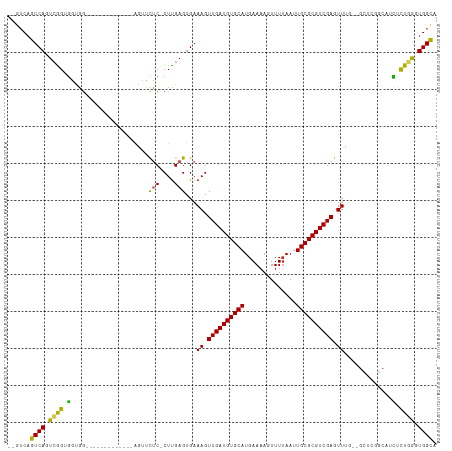
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:27 2011