| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,633,852 – 5,633,952 |
| Length | 100 |
| Max. P | 0.698288 |

| Location | 5,633,852 – 5,633,952 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 63.41 |
| Shannon entropy | 0.72540 |
| G+C content | 0.40897 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -7.85 |
| Energy contribution | -7.80 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.698288 |
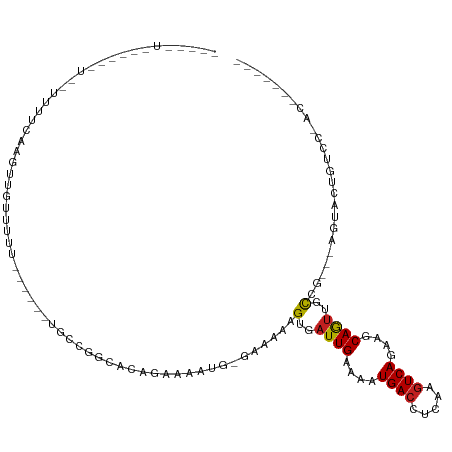
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5633852 100 - 23011544 -----UUUCCAGUUAUUUUCAAGUUGUUUUU------UGCCAGCGGAGAAAAUGGGAAAAAGUGAUUGAAAAUGACCUCAAGUCAGAAGCAGUUGCCG---AGUACUGUCCGAC------- -----......(((((((((((.(..(((((------(.(((..........))).))))))..)))))))))))).....(((.(..(((((.....---...)))))).)))------- ( -24.90, z-score = -1.53, R) >droGri2.scaffold_15252 3137785 89 - 17193109 ----------------UUUUGUGUUGUUUUCCAGAAACGCCGUGACAUAGAAUG-GAAAAAGUGAUUGAAAAUGACCUCAAGUCAAAAGCAAUUUCAGUCUCGUCC--------------- ----------------((((((((((((((...))))).....))))))))).(-((....(.((((((((.((((.....)))).......)))))))).).)))--------------- ( -15.80, z-score = -0.18, R) >droVir3.scaffold_12963 3604943 100 - 20206255 ----------------GCUCAUGUUGUUUUCAAGUUGCGCCGUGACACAAAAUG-GAAAAAGUGAUUGAAAAUGACCUCAAGUCAAAAGCAGUUUCAGUAGCCGGCAACUGUCCACC---- ----------------......((..(((((((.(..(.((((........)))-).....)..))))))))..))............((((((.(........).)))))).....---- ( -19.30, z-score = 0.63, R) >droMoj3.scaffold_6500 14003065 100 - 32352404 ----------------UUUCGUGUUGUUUUCUGGCUACCCUGUGACACAAAAUG-GAAAAAGUGAUUGAAAAUGACCUCAAGUCAAAAGCAGUUUCAGUAGCCAGCAACUGUCGUCC---- ----------------...((.((((....((((((((.((((........)))-).....(.(((((....((((.....))))....))))).).))))))))))))...))...---- ( -23.10, z-score = -1.02, R) >droWil1.scaffold_180708 3927306 85 + 12563649 -------------------UUUCUAGUUAUU------UUCCG---UAGAAAAUG-GAAAAAGUGAUUGAAAAUGACCUCAAGUCAAAAGCAGUUGCUGCGCAGAACUGUUCUGC------- -------------------..........((------(((((---(.....)))-)))))((..((((....((((.....))))....))))..))..(((((.....)))))------- ( -22.50, z-score = -2.59, R) >droPer1.super_10 1222616 96 + 3432795 UUUCCUUCUAGUUAUUUUCUAGGUUCCUUUA------UUGUGCCAUAGAAAAUG-GAAAAAGUGAUUGAAAAUGACCUCAAGUCAGCAGCAGUUGU-------GUCCGCC----------- .............(((((((((((.......------....))).))))))))(-((....(..((((....((((.....))))....))))..)-------.)))...----------- ( -21.10, z-score = -1.67, R) >dp4.chr4_group4 3395720 96 + 6586962 UUUCCUUCUAGUUAUUUUCUAGGUUCCUUUU------UUGUGCCAUAGAAAAUG-GAAAAAGUGAUUGAAAAUGACCUCAAGUCAGCAGCAGUUGU-------GUCCGCC----------- .............(((((((((((.......------....))).))))))))(-((....(..((((....((((.....))))....))))..)-------.)))...----------- ( -21.10, z-score = -1.54, R) >droAna3.scaffold_12916 11079021 92 - 16180835 -------------------UCUGCUGUUCUU------UGCCGGCAAAGAGAAUGAAAAAAAGUGAUUGAAAAUGAACUCAAGUCAGAAGCAGGUGCUG----GAACUGUCCUAGACCCAGC -------------------((((((.(((((------((....)))))))............(((((((........)).)))))..)))))).((((----(..(((...)))..))))) ( -22.70, z-score = -0.95, R) >droEre2.scaffold_4929 5722642 101 - 26641161 ---CAUCUGCAGUUAUUUUCAAGUU-GUUUU------UGCCAGCGCAGAAAAUGGGAAAAAGUGAUUGAAAAUGACCUCAAGUCAGAUGCAGUUGCCG---GGUACUGUCCCAC------- ---((.((((((((((((((((...-.((((------(.(((..........))).)))))....))))))))))).((......))))))).))..(---((......)))..------- ( -27.30, z-score = -1.33, R) >droYak2.chr2L 8764733 89 + 22324452 -----UCCCCAGUUAUUUUCAAGUUGGUUUU------UGCCAGCGGAGAAAAUGGGAAAAAGUGAUUGAAAAUGACCUCAAGUCAGAAGCAGUUGCCG---AC------------------ -----..((((....(((((..((((((...------.)))))).)))))..)))).....(..((((....((((.....))))....))))..)..---..------------------ ( -26.80, z-score = -3.06, R) >droSec1.super_5 3711648 100 - 5866729 -----UUUCAAGUUAUUUUCAAGUUGUUUUU------UGCCAGCGGAGAAAAUGGGAAAAAGUGAUUGAAAAUGACCUCAAGUCAGAAGCAGUUGCCG---AGUACUGUCCGAC------- -----......(((((((((((.(..(((((------(.(((..........))).))))))..)))))))))))).....(((.(..(((((.....---...)))))).)))------- ( -24.90, z-score = -1.54, R) >droSim1.chr2L 5426990 100 - 22036055 -----UUUCCAGUUAUUUUCAAGUUGUUUUU------UGCCAGCGGAGAAAAUGGGAAAAAGUGAUUGAAAAUGACCUCAAGUCAGAAGCAGUUGCCG---AGUACUGUCCAAC------- -----......(((((((((((.(..(((((------(.(((..........))).))))))..))))))))))))............(((((.....---...))))).....------- ( -23.70, z-score = -1.26, R) >consensus _____U______U__UUUUCAAGUUGUUUUU______UGCCGGCACAGAAAAUG_GAAAAAGUGAUUGAAAAUGACCUCAAGUCAGAAGCAGUUGCCG___AGUACUGUCC_AC_______ .............................................................(..((((....((((.....))))....))))..)......................... ( -7.85 = -7.80 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:32 2011