
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,254,066 – 18,254,157 |
| Length | 91 |
| Max. P | 0.739414 |

| Location | 18,254,066 – 18,254,157 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 74.30 |
| Shannon entropy | 0.51046 |
| G+C content | 0.37053 |
| Mean single sequence MFE | -15.10 |
| Consensus MFE | -7.16 |
| Energy contribution | -7.07 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18254066 91 + 24543557 CCCGUCUAAGGUA-GGGAAACCUUUCUC-UCAAUC----UCAACUUUUGCGCCUGUGUCAUCUAAUUGACAUUUGUUAAUAG-UUUCUUUUUCCAUUU-------- ........(((.(-((....))).))).-......----..((((.(((.((..((((((......))))))..))))).))-)).............-------- ( -16.40, z-score = -1.94, R) >droGri2.scaffold_15110 22164090 87 + 24565398 ------UGCAGCAUUCAGUGCCUUUCUC-UCGCUCAACUGCAUGCAUUGCGCCUGUGUCAUCUAAUUGACAUUUGUUAAUAGGUUUCUCCAUCU------------ ------.(((((((.((((.........-.......)))).))))..)))((..((((((......))))))..)).....((.....))....------------ ( -18.39, z-score = -0.96, R) >droWil1.scaffold_180949 733678 91 + 6375548 ---------------UUUUGUUUUUGUCGUCAUUAAAAUUAACCUUUUGCGCCUAUGUCAUCUAAUUGACAUUUGUUAAUAGUUAUUUUUUUCUCUUCUAUGAUUU ---------------.............(((((.((((.((((..((.(((...((((((......)))))).))).))..)))).)))).........))))).. ( -11.80, z-score = -2.45, R) >droPer1.super_20 1024407 94 + 1872136 ------CCCUGCAUACCGUGCCUUUCUC-UCAAUC---UCAAUCUUUUGCGCCUGUGUCAUCUAAUUGACAUUUGUUAAUAG-UUUUUCCCUCUAUUUUCUAUUU- ------....((((...)))).......-......---............((..((((((......))))))..)).(((((-.........)))))........- ( -8.90, z-score = -1.80, R) >dp4.chrXR_group6 11124807 94 + 13314419 ------CCCUGCAUACCAUGCCUUUCUC-UCAAUC---UCAAUCUUUUGCGCCUGUGUCAUCUAAUUGACAUUUGUUAAUAG-UUUUUCCCUCUAUUUUCUAUUU- ------....((((...)))).......-......---............((..((((((......))))))..)).(((((-.........)))))........- ( -8.60, z-score = -1.73, R) >droAna3.scaffold_13337 4160755 92 - 23293914 CCCGAUGAAGGUAUAGGAGACCUUUCUC-UCAAUC----UAAACUUUUGCGCCUGUGUCAUCUAAUUGACAUUUGUUAAUAG-UUUUUCUUUUCAGUU-------- ...((.((((((.......)))))).))-......----.(((((.(((.((..((((((......))))))..))))).))-)))............-------- ( -16.90, z-score = -0.91, R) >droEre2.scaffold_4784 10126939 91 - 25762168 CCCGGCUAAGGUA-GGGAAACCUUUCUC-UCAAUC----UCAACUUUUGCGCCUGUGUCAUCUAAUUGACAUUUGUUAAUAG-UUUUUCUUUCCAUUU-------- ...((..((((.(-((....))).....-......----..((((.(((.((..((((((......))))))..))))).))-))..)))).))....-------- ( -17.80, z-score = -1.70, R) >droYak2.chr3L 8407838 91 - 24197627 CCCGGCCAAGGUA-GGGAAACCUUUCUC-UCAAUC----UCAACUUUUGCGCCUGUGUCAUCUAAUUGACAUUUGUUAAUAG-UUUUUCUUUCCAUUU-------- ...((...(((.(-((....))).))).-......----..((((.(((.((..((((((......))))))..))))).))-)).......))....-------- ( -17.60, z-score = -1.47, R) >droSec1.super_19 248088 91 + 1257711 CCCGGCUAUGGUA-GGGAAACCUUUCUC-UCAAUC----UCAACUUUUGCGCCUGUGUCAUCUAAUUGACAUUUGUUAAUAG-UUUUUCUUUCCAUUU-------- ...((....((.(-((....))).))..-......----..((((.(((.((..((((((......))))))..))))).))-)).......))....-------- ( -16.80, z-score = -1.46, R) >droSim1.chr3L 17583680 91 + 22553184 CCCGGCUAAGGUA-GGGAAACCUUUCUC-UCAAUC----UCAACUUUUGCGCCUGUGUCAUCUAAUUGACAUUUGUUAAUAG-UUUUUCUUUCCAUUU-------- ...((..((((.(-((....))).....-......----..((((.(((.((..((((((......))))))..))))).))-))..)))).))....-------- ( -17.80, z-score = -1.70, R) >consensus CCCG_CUAAGGUA_GGGAAACCUUUCUC_UCAAUC____UCAACUUUUGCGCCUGUGUCAUCUAAUUGACAUUUGUUAAUAG_UUUUUCUUUCCAUUU________ ..................................................((..((((((......))))))..)).............................. ( -7.16 = -7.07 + -0.09)
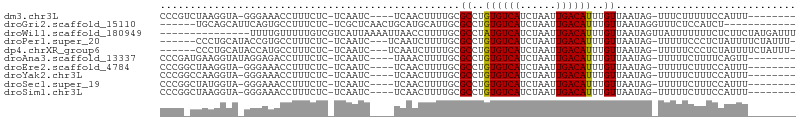

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:23 2011