| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,181,120 – 18,181,216 |
| Length | 96 |
| Max. P | 0.785859 |

| Location | 18,181,120 – 18,181,216 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 70.03 |
| Shannon entropy | 0.56405 |
| G+C content | 0.37987 |
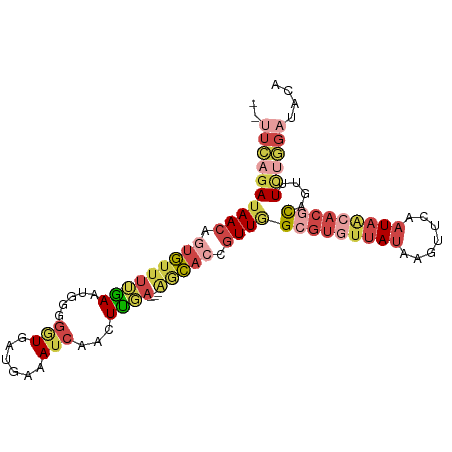
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -10.45 |
| Energy contribution | -12.65 |
| Covariance contribution | 2.20 |
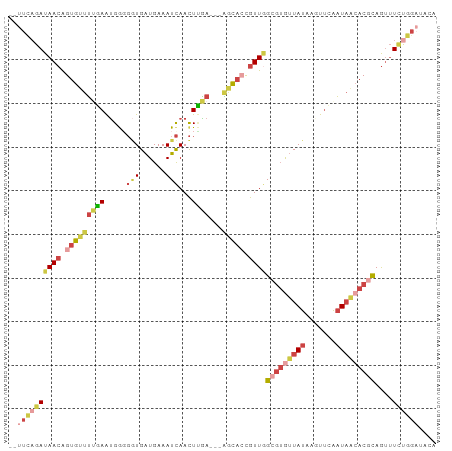
| Combinations/Pair | 1.37 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
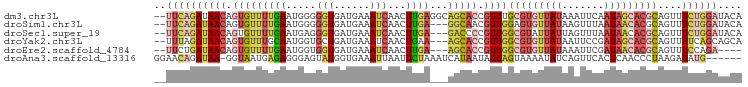
>dm3.chr3L 18181120 96 + 24543557 --UUCAGAUAACAGUGUUUUGAAUGGGGGUGAUGAAAUCAACUUGAGGCAGCACCGUUUGCGUGUUAUAAAUUCAAUAGCACGCAGUUUCUGGAUACA --((((((.(((...............((((.((...((.....))..)).))))...((((((((((.......))))))))))))))))))).... ( -27.90, z-score = -2.26, R) >droSim1.chr3L 17511394 93 + 22553184 --UUCAGAUAACAGUGUUUUGAAUGGGGGUGAUGAAAUCAACUUGA---GGCAACGUUGGAGUGUUAUAAGUUUAAUAACACGCAGUUUCUGGAUACA --((((((((((..(((((..(..(..(((......)))..))..)---))))..))))..(((((((.......)))))))......)))))).... ( -20.90, z-score = -1.18, R) >droSec1.super_19 176189 93 + 1257711 --UUCAGAUAACAGUGUUUUGAAUGAGGGUGAUGAAAUCAACUUGA---GACCCCGUUGGCGUAUUAUUAGUUUAAUAACACGCAGUUUCUGGAUACA --((((((.(((.(((((((((((...(((((((...(((((....---......)))))..))))))).)))))).)))))...))))))))).... ( -21.50, z-score = -1.18, R) >droYak2.chr3L 8331169 93 - 24197627 --UUUAGAUAACAGUGUUUGCAAUGGUGCUGAUGAAAUCAACUGAA---AGCACCGUUGGCGUGUUAUAAUUCCGAUAGCACGCAGUUUUCAGCAGCA --....((.(((........((((((((((................---))))))))))(((((((((.......))))))))).))).))....... ( -29.99, z-score = -2.65, R) >droEre2.scaffold_4784 10053755 89 - 25762168 --UUCUGAUAACAGUGUUUUGAAUGGUGGUGAUGAAAUCAACUUGA---AGCACCGUUGGCGUGUUAUAAAUUCGAUAACACGCAGUUUCCAGA---- --.((((.((((.((((((..(....((((......))))..)..)---))))).))))(((((((((.......)))))))))......))))---- ( -28.30, z-score = -3.21, R) >droAna3.scaffold_13316 949 91 - 1404 GGAACAGAUAA-GGUAAUGAGAGGGAGUAUGGUGAAAUUAAUUCUAAAUCAUAAUAUUAGUAAAAUAUCAGUUCACUCAACCCUAAGAUAUG------ ..........(-((...((((.(((....(((((..((((((..((.....))..))))))....))))).))).))))..)))........------ ( -9.70, z-score = 1.43, R) >consensus __UUCAGAUAACAGUGUUUUGAAUGGGGGUGAUGAAAUCAACUUGA___AGCACCGUUGGCGUGUUAUAAGUUCAAUAACACGCAGUUUCUGGAUACA ..((((((((((.(((((......((...((((...)))).))......))))).))))(((((((((.......)))))))))....)))))).... (-10.45 = -12.65 + 2.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:20 2011