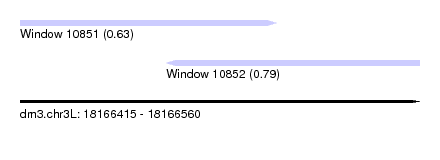
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,166,415 – 18,166,560 |
| Length | 145 |
| Max. P | 0.793891 |

| Location | 18,166,415 – 18,166,508 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 97.13 |
| Shannon entropy | 0.03950 |
| G+C content | 0.63441 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -35.57 |
| Energy contribution | -35.47 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.634344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
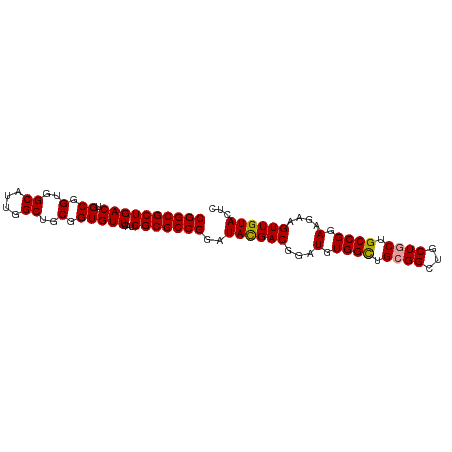
>dm3.chr3L 18166415 93 + 24543557 CGGCGCUGACUGCGGUGGCAUUGGCUGCGGUGUUGAUGGCGCCGGAUAUGACGGAUGUGGUUGCGGCUGCUGCUGCCGGAAGAAGUUGUACUC ((((((((((.((.(..((....))..).)))))...)))))))..((..((..........(((((....)))))(....)..))..))... ( -34.80, z-score = -0.67, R) >droSec1.super_19 161965 93 + 1257711 CGGCGCUGACUGCGGUGGCAUUGGCUGCGGUGUUGAUGGCGCCGGAUACGACGGAUGUGGCUGCGGCUGCUGCUGCCGGAAGAAGUUGUAUUC .(((((((((.((.(..((....))..).)))))...))))))(((((((((...(.((((.((((...)))).)))).)....))))))))) ( -40.60, z-score = -2.20, R) >droSim1.chr3L 17494014 93 + 22553184 CGGCGCUGACUGCGGUGGCAUUGGCUGCGGUGUUGAUGGCGCCGGAUACGACGGAUGUGGCUGGGGCUGCUGCUGCCGGAAGAAGUUGUACUC ((((((((((.((.(..((....))..).)))))...)))))))..((((((.....(..(((((((....))).))))..)..))))))... ( -35.90, z-score = -0.81, R) >consensus CGGCGCUGACUGCGGUGGCAUUGGCUGCGGUGUUGAUGGCGCCGGAUACGACGGAUGUGGCUGCGGCUGCUGCUGCCGGAAGAAGUUGUACUC ((((((((((.((.(..((....))..).)))))...)))))))..((((((...(.((((.((((...)))).)))).)....))))))... (-35.57 = -35.47 + -0.11)

| Location | 18,166,468 – 18,166,560 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 78.27 |
| Shannon entropy | 0.34885 |
| G+C content | 0.65723 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.12 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.793891 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
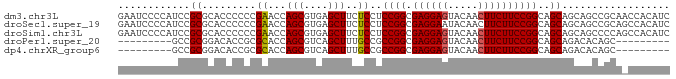
>dm3.chr3L 18166468 92 - 24543557 GAAUCCCCAUCCGCGCACCCCCCGAACCAGCGUGAGCUUCUCCUCCGGCGAGGAGUACAACUUCUUCCGGCAGCAGCAGCCGCAACCACAUC ...........(((((.............))))).(((.((.((((((.((((((.....)))))))))).)).)).)))............ ( -24.22, z-score = -1.39, R) >droSec1.super_19 162018 92 - 1257711 GAAUCCCCAUCCGCGCACCCCCCGAACCAGCGUGAGCUUCUCCUCCGGCGAGGAAUACAACUUCUUCCGGCAGCAGCAGCCGCAGCCACAUC ...........(((((.............)))))............(((((((((......))))))((((.......))))..)))..... ( -21.62, z-score = -0.32, R) >droSim1.chr3L 17494067 92 - 22553184 GAAUCCCCAUCCGCGCACCCCCCGAACCAGCGUGAGCUUCUCCUCCGGCGAGGAGUACAACUUCUUCCGGCAGCAGCAGCCCCAGCCACAUC ...........(((((.............))))).(((.((.((((((.((((((.....)))))))))).)).)).)))............ ( -24.22, z-score = -1.45, R) >droPer1.super_20 926329 74 - 1872136 ---------GCCGCGGACACCGCGCACCAGCGUCAGCUUUGCCGCCGGCGAGGAGUACAACUUCUUCCGGCAGCAGACACAGC--------- ---------((.((((...))))))....((....))(((((.(((((.((((((.....))))))))))).)))))......--------- ( -30.30, z-score = -2.48, R) >dp4.chrXR_group6 11027244 74 - 13314419 ---------GCCGCGGACACCGCGCACCAGCGUCAGCUUUGCCGCCGGCGAGGAGUACAACUUCUUCCGGCAGCAGACACAGC--------- ---------((.((((...))))))....((....))(((((.(((((.((((((.....))))))))))).)))))......--------- ( -30.30, z-score = -2.48, R) >consensus GAAUCCCCAUCCGCGCACCCCCCGAACCAGCGUGAGCUUCUCCUCCGGCGAGGAGUACAACUUCUUCCGGCAGCAGCAGCCGCA_CCACAUC ............((.........((...(((....)))..))..((((.((((((.....))))))))))..)).................. (-18.40 = -18.12 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:19 2011