| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,147,829 – 18,147,928 |
| Length | 99 |
| Max. P | 0.938623 |

| Location | 18,147,829 – 18,147,928 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Shannon entropy | 0.40150 |
| G+C content | 0.37260 |
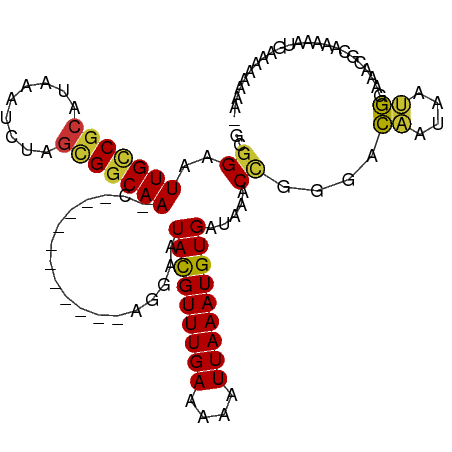
| Mean single sequence MFE | -20.28 |
| Consensus MFE | -14.22 |
| Energy contribution | -13.69 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
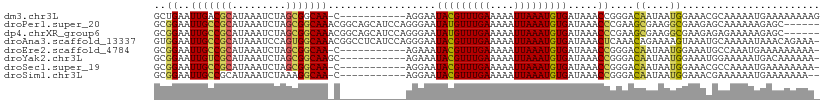
>dm3.chr3L 18147829 99 - 24543557 GCUGAAUUGACGCAUAAAUCUAGCGGCAA-C-----------AGGAAUACGUUUGAAAAAUUAAAUGUGAUAAACCGGGACAAUAAUGGAAACGCAAAAAUGAAAAAAAAG ((((.(((........))).)))).((..-.-----------.((..(((((((((....))))))))).....))...........(....)))................ ( -13.90, z-score = -1.09, R) >droPer1.super_20 902589 105 - 1872136 GCGGAAUUGCCGCAUAAAUCUAGCGGCAAACGGCAGCAUCCAGGGAAUAUGUUUGAAAAAUUAAAUGUGAUAAACCCGAAGCGAAGGCGAAGAGCAAAAAAGAGC------ ((.(..(((((((.........))))))).).)).((.....(((..(((((((((....))))))))).....)))...((....)).....))..........------ ( -26.70, z-score = -1.69, R) >dp4.chrXR_group6 10991370 105 - 13314419 GCGGAAUUGCCGCAUAAAUCUAGCGGCAAACGGCAGCAUCCAGGGAAUAUGUUUGAAAAAUUAAAUGUGAUAAACCCGAAGCGAAGGCGAAGAGAGAAAAAGAGC------ ((.(..(((((((.........))))))).).)).((.....(((..(((((((((....))))))))).....)))...((....))...............))------ ( -25.00, z-score = -1.51, R) >droAna3.scaffold_13337 4062279 110 + 23293914 GUGGAAUUGCCGCAUAAAUCCAGUGGCAAACGGCCUCAUCCAGGGAAUACGUUUGAAAAAUUAAAUGUGAUAAACUCAAACAGAAAAGUAAAUGCAAAAAUAAACAGAAA- (.((..(((((((.........)))))))....)).).....(((..(((((((((....))))))))).....))).................................- ( -17.30, z-score = -0.07, R) >droEre2.scaffold_4784 10020944 98 + 25762168 GCGGAAUUGCCGCAUAAAUCUAGCGGCAA-C-----------AGAAAUACGUUUGAAAAAUUAAAUGUGAUAAACCGGGACAAUAAUGGAAAUGCCAAAUGAAAAAAAAA- .(((..(((((((.........)))))))-.-----------.....(((((((((....))))))))).....))).........(((.....))).............- ( -22.20, z-score = -3.01, R) >droYak2.chr3L 8297326 99 + 24197627 GCGGAAUUGUCGCAUAAAUCUAGCGGCAAGC-----------AGAAAUACGUUUGAAAAAUUAAAUGUGAUAAACCGGGACAAUAAUGGAAAUGGAAAAAUGACAAAAAA- .(((..(((((((.........)))))))..-----------.....(((((((((....))))))))).....))).................................- ( -18.10, z-score = -1.94, R) >droSec1.super_19 143658 98 - 1257711 GCGGAAUUGCCGCAUAAAUCUAGCGGCAA-C-----------AGGAAUACGUUUGAAAAAUUAAAUGUGAUAAACCGGGACAAUAAUGGAAACGCCAAAAUGAAAAAAAA- .(((..(((((((.........)))))))-.-----------.....(((((((((....))))))))).....))).........(((.....))).............- ( -22.20, z-score = -2.63, R) >droSim1.chr3L 17475846 97 - 22553184 GCGGAAUUGCCGCAUAAAUCUAAAGGCAA-C-----------AGGAAUACGUUUGAAAAAUUAAAUGUGAUAAACCGGGACAAUAAUGGAAACGAAAAAAUGAAAAAAA-- .(((..(((((.............)))))-.-----------.....(((((((((....))))))))).....))).........((....))...............-- ( -16.82, z-score = -2.19, R) >consensus GCGGAAUUGCCGCAUAAAUCUAGCGGCAA_C___________AGGAAUACGUUUGAAAAAUUAAAUGUGAUAAACCGGGACAAUAAUGGAAACGCAAAAAUGAAAAAAAA_ ..((..(((((((.........)))))))..................(((((((((....))))))))).....))....((....))....................... (-14.22 = -13.69 + -0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:18 2011