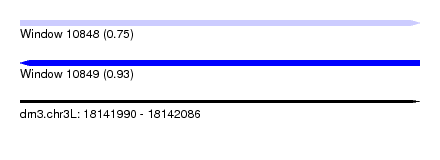
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,141,990 – 18,142,086 |
| Length | 96 |
| Max. P | 0.925485 |

| Location | 18,141,990 – 18,142,086 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 68.79 |
| Shannon entropy | 0.66634 |
| G+C content | 0.56434 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -13.81 |
| Energy contribution | -14.40 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18141990 96 + 24543557 UGCUUUUGCGGCUUAAAGCGGCAGCAGGUCGCUUCAGUCAGUCGAAGACUUUAAGCGCACCAAGUCGCCU------GCCGCUCCAC---UCCAAAAAAUACGAGA .((....))((.....(((((((((.((((((((.((((.......))))..))))).))).....).))------))))))....---.))............. ( -29.50, z-score = -0.89, R) >droMoj3.scaffold_6680 15725909 73 + 24764193 CGCUUUGGCGGCCCGCUGCGGCAUUAACUUGGCUCAGUCACUCGAAGACUUUAAACGCACCAAGUUGCCUCGU-------------------------------- .......((((....))))((((...((((((..(((((.......))))......)..))))))))))....-------------------------------- ( -18.50, z-score = 0.57, R) >droVir3.scaffold_13049 21319206 98 + 25233164 CGCUUUGGCGGCUUGAGGCGGCCCCAAGUCGCGUCAGUCAUUCGAAGACUUUAAACGCACCAAGUUGCCUAGUUCGGCUGCACGACUACUCGACUCCU------- ......(((((((((..(((.....(((((...((........)).)))))....)))..)))))))))((((.((......))))))..........------- ( -30.10, z-score = -0.27, R) >droGri2.scaffold_15110 2252827 81 - 24565398 CGCUUUGGCGGCUCGAAGCGGACCCAAGUCGCUUCAGUCACUCAAAGACCUGAAGCGCACCAAGUUGUUGCCUCGGCUUCC------------------------ .(((..((((((.(...((.(((....)))(((((((((.......)).))))))))).....)..))))))..)))....------------------------ ( -27.40, z-score = -0.98, R) >droWil1.scaffold_180949 5452835 90 - 6375548 CGCUUUGGCGGCUCCAAACGGCACCAAGUCGCUUCAGUCAGUCAAAGACUUUAAACGCAUCAAGUUGGCCAGUUAGUUGGAGCUGUUGAU--------------- ....(..(((((((((((((((...((((((((......)))....)))))..(((.......))).))).))...))))))))))..).--------------- ( -26.40, z-score = -0.97, R) >droAna3.scaffold_13337 4057071 98 - 23293914 CGCUUUGGCGGCUUUAAGCGGCUGCAGGUCGCUUCAGUCAGUCCAAGACUUUAAGCGCACCAAGUCGCCA------GCCGCUCCGUGGAUCCGGUGGAUGUGGU- (((...(((((((....((((((...((((((((.((((.......))))..))))).))).)))))).)------))))))..))).((((...)))).....- ( -37.70, z-score = -0.90, R) >droEre2.scaffold_4784 10014937 95 - 25762168 CGCUUUGGCGGCUUUAAGCGGCAGCAGGUCGCUUCAGUCAGUCCAAGACUUUAAGCGCACCAAGUCGCCU------GCCGCUCCAC---UCGAAAAG-GCCAAGA .(((((..(((.....(((((((((.((((((((.((((.......))))..))))).))).....).))------))))))....---)))..)))-))..... ( -33.90, z-score = -1.30, R) >droYak2.chr3L 8291497 102 - 24197627 CGCUUUGGCGGCUUUAAGCGGCCGCAGGUCGCUUCAGUCAGUCCAAGACUUUAAGCGCACCAAGUCGCCUUCGCCUGCAGCUCCAC---UCAAAAAAAGCCGAGA .(((((((((((((...((....)).((((((((.((((.......))))..))))).)))))))))))...((.....)).....---......)))))..... ( -32.30, z-score = -0.54, R) >droSec1.super_19 137854 95 + 1257711 CGCUUUUGCGGCUUAAAGCGGCAGCAGGUCGCUUCAGUCAGUCCAAGACUUUAAGCGCACCAAGUCGCCU------GCCGCUCCAC---UCCAAAAA-GCCGAGA .((((((..((.....(((((((((.((((((((.((((.......))))..))))).))).....).))------))))))....---.)).))))-))..... ( -33.90, z-score = -2.21, R) >droSim1.chr3L 17469967 95 + 22553184 CGCUUUUGCGGCUUAAAGCGGCAGCAGGUCGCUUCAGUCAGUCCAAGACUUUAAGCGCACCAAGUCGCCU------GCCGCUCCAC---UCCAAAAA-GCCGAGU .((((((..((.....(((((((((.((((((((.((((.......))))..))))).))).....).))------))))))....---.)).))))-))..... ( -33.90, z-score = -2.53, R) >consensus CGCUUUGGCGGCUUAAAGCGGCAGCAGGUCGCUUCAGUCAGUCCAAGACUUUAAGCGCACCAAGUCGCCU______GCCGCUCCAC___UCCAAAAA___CGAG_ .......((((((..(((((((.....))))))).((((.......))))............))))))..................................... (-13.81 = -14.40 + 0.59)

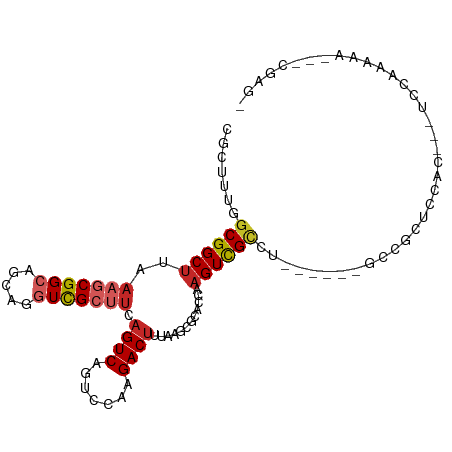
| Location | 18,141,990 – 18,142,086 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 68.79 |
| Shannon entropy | 0.66634 |
| G+C content | 0.56434 |
| Mean single sequence MFE | -34.71 |
| Consensus MFE | -15.17 |
| Energy contribution | -15.53 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.925485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
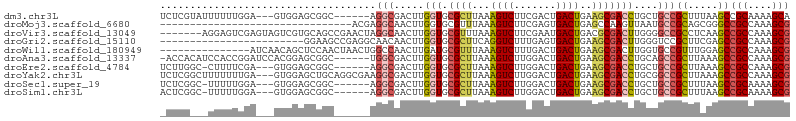
>dm3.chr3L 18141990 96 - 24543557 UCUCGUAUUUUUUGGA---GUGGAGCGGC------AGGCGACUUGGUGCGCUUAAAGUCUUCGACUGACUGAAGCGACCUGCUGCCGCUUUAAGCCGCAAAAGCA ....((.((((.(((.---.(((((((((------((.((....(((.(((((..((((.......)))).)))))))))))))))))))))..))).)))))). ( -38.50, z-score = -2.22, R) >droMoj3.scaffold_6680 15725909 73 - 24764193 --------------------------------ACGAGGCAACUUGGUGCGUUUAAAGUCUUCGAGUGACUGAGCCAAGUUAAUGCCGCAGCGGGCCGCCAAAGCG --------------------------------....(((((((((((........((((.......))))..))))))))...(((......))).)))...... ( -22.30, z-score = -0.01, R) >droVir3.scaffold_13049 21319206 98 - 25233164 -------AGGAGUCGAGUAGUCGUGCAGCCGAACUAGGCAACUUGGUGCGUUUAAAGUCUUCGAAUGACUGACGCGACUUGGGGCCGCCUCAAGCCGCCAAAGCG -------......(((....))).((.(((......)))...((((((((((...((((.......)))))))))(.(((((((...))))))).)))))).)). ( -30.50, z-score = 0.50, R) >droGri2.scaffold_15110 2252827 81 + 24565398 ------------------------GGAAGCCGAGGCAACAACUUGGUGCGCUUCAGGUCUUUGAGUGACUGAAGCGACUUGGGUCCGCUUCGAGCCGCCAAAGCG ------------------------((..((((((((....(((..(..((((((((.((.......))))))))))..)..)))..)))))).))..))...... ( -32.50, z-score = -1.52, R) >droWil1.scaffold_180949 5452835 90 + 6375548 ---------------AUCAACAGCUCCAACUAACUGGCCAACUUGAUGCGUUUAAAGUCUUUGACUGACUGAAGCGACUUGGUGCCGUUUGGAGCCGCCAAAGCG ---------------.......(((((((...((.(((..(((.....(((((..((((.......)))).)))))....)))))))))))))))(((....))) ( -25.20, z-score = -1.19, R) >droAna3.scaffold_13337 4057071 98 + 23293914 -ACCACAUCCACCGGAUCCACGGAGCGGC------UGGCGACUUGGUGCGCUUAAAGUCUUGGACUGACUGAAGCGACCUGCAGCCGCUUAAAGCCGCCAAAGCG -..........(((......))).(((((------(((((.((.(((.(((((..((((.......)))).))))))))...)).))))...))))))....... ( -35.10, z-score = -1.43, R) >droEre2.scaffold_4784 10014937 95 + 25762168 UCUUGGC-CUUUUCGA---GUGGAGCGGC------AGGCGACUUGGUGCGCUUAAAGUCUUGGACUGACUGAAGCGACCUGCUGCCGCUUAAAGCCGCCAAAGCG ..(((((-......(.---((.(((((((------((.((....(((.(((((..((((.......)))).)))))))))))))))))))...)))))))).... ( -35.30, z-score = -0.71, R) >droYak2.chr3L 8291497 102 + 24197627 UCUCGGCUUUUUUUGA---GUGGAGCUGCAGGCGAAGGCGACUUGGUGCGCUUAAAGUCUUGGACUGACUGAAGCGACCUGCGGCCGCUUAAAGCCGCCAAAGCG ....(((...((((((---(((..(((((((((.(((....))).)..(((((..((((.......)))).))))).)))))))))))))))))..)))...... ( -40.50, z-score = -1.21, R) >droSec1.super_19 137854 95 - 1257711 UCUCGGC-UUUUUGGA---GUGGAGCGGC------AGGCGACUUGGUGCGCUUAAAGUCUUGGACUGACUGAAGCGACCUGCUGCCGCUUUAAGCCGCAAAAGCG .....((-(((((((.---.(((((((((------((.((....(((.(((((..((((.......)))).)))))))))))))))))))))..))).)))))). ( -43.60, z-score = -3.16, R) >droSim1.chr3L 17469967 95 - 22553184 ACUCGGC-UUUUUGGA---GUGGAGCGGC------AGGCGACUUGGUGCGCUUAAAGUCUUGGACUGACUGAAGCGACCUGCUGCCGCUUUAAGCCGCAAAAGCG .....((-(((((((.---.(((((((((------((.((....(((.(((((..((((.......)))).)))))))))))))))))))))..))).)))))). ( -43.60, z-score = -3.29, R) >consensus _CUCG___UUUUUGGA___GUGGAGCGGC______AGGCGACUUGGUGCGCUUAAAGUCUUGGACUGACUGAAGCGACCUGCUGCCGCUUAAAGCCGCCAAAGCG ....................................(((.....(((.((((...((((.......))))..)))))))....)))((.....))(((....))) (-15.17 = -15.53 + 0.36)
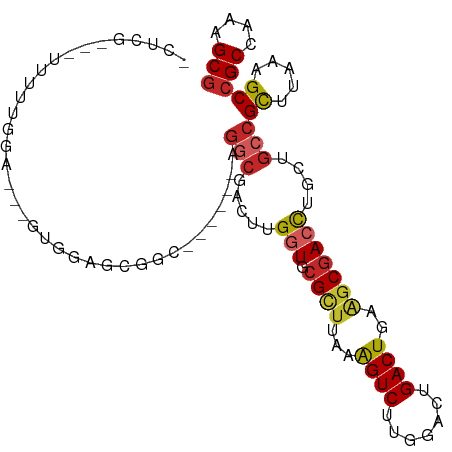
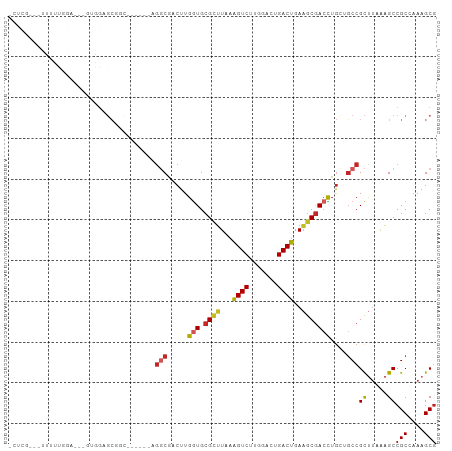
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:17 2011