| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,138,116 – 18,138,225 |
| Length | 109 |
| Max. P | 0.833950 |

| Location | 18,138,116 – 18,138,225 |
|---|---|
| Length | 109 |
| Sequences | 13 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.40 |
| Shannon entropy | 0.61401 |
| G+C content | 0.53368 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.52 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.833950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
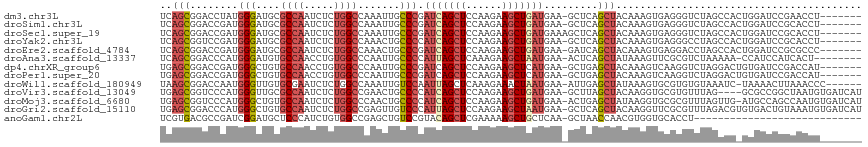
>dm3.chr3L 18138116 109 + 24543557 UCAGCGGACCUAUGGGAUGCGCCAAUCUCUGGCCAAAUUGCCCGAUCAGCUCCAAGAAGCUGAUGAA-GCUCAGCUACAAAGUGAGGGUCUAGCCACUGGAUCCGAACCU------- ..(((.((.((.((((.((.((((.....)))))).....))))(((((((......)))))))..)-).)).))).....((..((((((((...))))))))..))..------- ( -35.10, z-score = -0.81, R) >droSim1.chr3L 17465966 109 + 22553184 UCAGCGGACCGAUGGGAUGCGCCAAUCUCUGGCCAAAUUGCCCGAUCAGCUCCAAGAAGCUGAUGAA-GCUCAGCUACAAAGUGAGGGUCUAGCCACUGGAUCCGCACCU------- ...((((((((.(((..((.((((.....))))))....((((.(((((((......)))))))(.(-((...))).).......))))....))).))).)))))....------- ( -38.20, z-score = -1.28, R) >droSec1.super_19 133912 110 + 1257711 UCAGCGGACCGAUGGGAUGCGCCAAUCUCUGGCCAAAUUGCCCGAUCAGCUCCAAGAAGCUGAUGAAAGCUCAGCUACAAAGUGAGGGUCUAGCCACUGGAUCCGCACCU------- ...((((((((.(((..((.((((.....))))))....((((.(((((((......)))))))...(((...))).........))))....))).))).)))))....------- ( -38.10, z-score = -1.34, R) >droYak2.chr3L 8287289 109 - 24197627 UCAGCGGUCCGAUGGGAUGCGCCAAUCUCUGGCCAAACUGCCCCAUCAGCUCCAAGAAGCUGAUGAA-GCUCAGCUACAAAGUGAGGGCCUAGCCACUGGAUCCGCACCU------- ...((((((((.(((..((.((((.....))))))....((((((((((((......)))))))).(-((...))).........))))....))).)))).))))....------- ( -42.00, z-score = -2.11, R) >droEre2.scaffold_4784 10010981 109 - 25762168 UCAGCGGACCGAUGGGAUGCGCCAAUCUCUGGCCAAACUGCCCGAUCAGCUCCAAGAAGCUGAUGAA-GAUCAGCUACAAAGUGAGGACCUAGCCACUGGAUCCGCGCCC------- ...(((((((((((((.((.((((.....)))))).....))).)))((.(((....(((((((...-.))))))).(.....).))).)).......)).)))))....------- ( -35.40, z-score = -1.13, R) >droAna3.scaffold_13337 4053475 107 - 23293914 UCAGCGGACCCAUGGGAUGUGCCAACCUGUGGCCCAAUUGCCCCAUUAGCUCAAAGAAGCUAAUGAA-ACUCAGCUAUAAAGUUCGCGUCUAAAAA-CCAUCCAUCACU-------- ...((((((....(((.((.((((.....)))).))....)))((((((((......))))))))..-.............)))))).........-............-------- ( -27.90, z-score = -2.24, R) >dp4.chrXR_group6 10980001 109 + 13314419 UGAGCGGACCGAUGGGCUGUGCCAACCUGUGGCCCAAUUGCCCGAUCAGCUCCAAGAAGCUCAUGAA-GCUGAGCUACAAAGUCAAGGUCUAGGACUGUGAUCCGACCAU------- (((..((((.(((((((((.((((.....)))).))...)))).))).).)))..(.((((((....-..)))))).)....))).((((..(((......)))))))..------- ( -34.80, z-score = -0.90, R) >droPer1.super_20 890694 109 + 1872136 UGAGCGGACCGAUGGGCUGUGCCAACCUGUGGCCCAAUUGCCCGAUCAGCUCCAAGAAGCUCAUGAA-GCUGAGCUACAAAGUCAAGGUCUAGGACUGUGAUCCGACCAU------- (((..((((.(((((((((.((((.....)))).))...)))).))).).)))..(.((((((....-..)))))).)....))).((((..(((......)))))))..------- ( -34.80, z-score = -0.90, R) >droWil1.scaffold_180949 5446580 109 - 6375548 UAAGCGGACCAAUGGGUUGUGCGAAUCUCUGGCCAAAUUGUCCAAUUAGCUCAAAGAAACUAAUGAA-AUUGAGCUAUAAAGUGCGUGUGUAAAUC-UAAAACUUAAACCC------ ...((((((.....((((............)))).....))))...((((((((.............-.))))))))......))...........-..............------ ( -19.24, z-score = -0.14, R) >droVir3.scaffold_13049 21314329 112 + 25233164 UGAGCGGUCCCAUGGGUUGCGCCAAUCUCUGGCCGAACUGCCCCAUCAGCUCCAAGAAGCUGAUGAA-GCUUAGCUACAAGGUGCGUGUUUAG----GCGCCGGCUAAUGUGAUCAU .....((((.(((((((..(((((.....))).))....))))((((((((......))))))))..-...(((((....(((((........----))))))))))))).)))).. ( -39.20, z-score = -0.71, R) >droMoj3.scaffold_6680 15720548 115 + 24764193 UGAGCGGUCCCAUGGGCUGUGCCAAUCUCUGGCCCAACUGCCCCAUCAGCUCCAAGAAGCUGAUGAA-ACUGAGCUAUAAGGUGCGCGUUUAGUUG-AUGCCAGCCAAUGUGAUCAU .....((((.(((.(((((.((((.....)))).(((((....((((((((......))))))))((-((...((........))..)))))))))-....))))).))).)))).. ( -39.80, z-score = -1.76, R) >droGri2.scaffold_15110 2246786 116 - 24565398 UGAGCGGACCCAUGGGCUGUGCCAAUCUCUGGCCGAGUUGUCCCAUUAGCUCCAAGAAGCUAAUGAA-GCUCAGCUACAAGGUUCGCGUUUAGACGUGUGACUGUAAAUGUGAUCAU ...(((((((....(((((.((((.....)))))(((((....((((((((......)))))))).)-))))))))....))))))).....((((..(........)..)).)).. ( -39.00, z-score = -2.08, R) >anoGam1.chr2L 10631140 88 + 48795086 UCGUGACGCCGAUCGGAUGCUCCCAUCUGUGGCCGAGCUGUCCGUACAGCUCGAAAAAGCUGCUCAA-GCUAACCAACGUGGUGCACCU---------------------------- ..(((.(((((..((((((....))))))(((.((((((((....))))))))....((((.....)-)))..)))...))))))))..---------------------------- ( -33.00, z-score = -2.89, R) >consensus UCAGCGGACCGAUGGGAUGCGCCAAUCUCUGGCCAAAUUGCCCGAUCAGCUCCAAGAAGCUGAUGAA_GCUCAGCUACAAAGUGAGGGUCUAGCCA_UGGAUCCGCACCU_______ ..(((........(((....((((.....)))).......))).(((((((......))))))).........)))......................................... (-15.28 = -15.52 + 0.24)
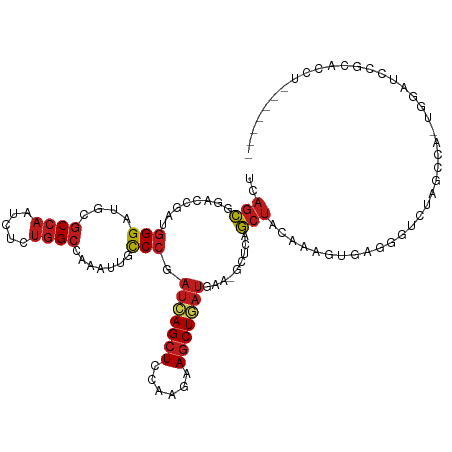

| Location | 18,138,116 – 18,138,225 |
|---|---|
| Length | 109 |
| Sequences | 13 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.40 |
| Shannon entropy | 0.61401 |
| G+C content | 0.53368 |
| Mean single sequence MFE | -38.11 |
| Consensus MFE | -16.52 |
| Energy contribution | -16.35 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.583155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
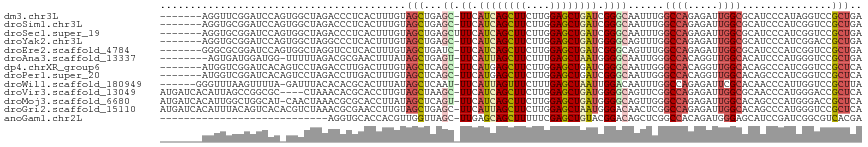
>dm3.chr3L 18138116 109 - 24543557 -------AGGUUCGGAUCCAGUGGCUAGACCCUCACUUUGUAGCUGAGC-UUCAUCAGCUUCUUGGAGCUGAUCGGGCAAUUUGGCCAGAGAUUGGCGCAUCCCAUAGGUCCGCUGA -------.....((((.((.((((......((...(((((..((..(((-(..((((((((....))))))))..)))...)..)))))))...))......)))).)))))).... ( -36.90, z-score = -0.05, R) >droSim1.chr3L 17465966 109 - 22553184 -------AGGUGCGGAUCCAGUGGCUAGACCCUCACUUUGUAGCUGAGC-UUCAUCAGCUUCUUGGAGCUGAUCGGGCAAUUUGGCCAGAGAUUGGCGCAUCCCAUCGGUCCGCUGA -------....(((((((...((((((((...(((((....)).)))((-(..((((((((....))))))))..)))..))))))))..(((.((.....)).))))))))))... ( -42.20, z-score = -1.11, R) >droSec1.super_19 133912 110 - 1257711 -------AGGUGCGGAUCCAGUGGCUAGACCCUCACUUUGUAGCUGAGCUUUCAUCAGCUUCUUGGAGCUGAUCGGGCAAUUUGGCCAGAGAUUGGCGCAUCCCAUCGGUCCGCUGA -------....(((((((...((((((((...(((((....)).)))((((..((((((((....)))))))).))))..))))))))..(((.((.....)).))))))))))... ( -40.30, z-score = -0.59, R) >droYak2.chr3L 8287289 109 + 24197627 -------AGGUGCGGAUCCAGUGGCUAGGCCCUCACUUUGUAGCUGAGC-UUCAUCAGCUUCUUGGAGCUGAUGGGGCAGUUUGGCCAGAGAUUGGCGCAUCCCAUCGGACCGCUGA -------....((((.(((.((((....((((...(((((..((..(((-(((((((((((....)))))))))))))...)..)))))))...)).))...)))).)))))))... ( -50.10, z-score = -2.42, R) >droEre2.scaffold_4784 10010981 109 + 25762168 -------GGGCGCGGAUCCAGUGGCUAGGUCCUCACUUUGUAGCUGAUC-UUCAUCAGCUUCUUGGAGCUGAUCGGGCAGUUUGGCCAGAGAUUGGCGCAUCCCAUCGGUCCGCUGA -------....(((((.((.((((..((((....))))(((.((..(((-((.((((((((....))))))))..(((......))).)))))..)))))..)))).)))))))... ( -42.20, z-score = -0.67, R) >droAna3.scaffold_13337 4053475 107 + 23293914 --------AGUGAUGGAUGG-UUUUUAGACGCGAACUUUAUAGCUGAGU-UUCAUUAGCUUCUUUGAGCUAAUGGGGCAAUUGGGCCACAGGUUGGCACAUCCCAUGGGUCCGCUGA --------((((((((((((-((....)))((.(((((....(((.(((-..(((((((((....)))))))))..))...).)))...))))).)).))))).....)).)))).. ( -33.70, z-score = -1.08, R) >dp4.chrXR_group6 10980001 109 - 13314419 -------AUGGUCGGAUCACAGUCCUAGACCUUGACUUUGUAGCUCAGC-UUCAUGAGCUUCUUGGAGCUGAUCGGGCAAUUGGGCCACAGGUUGGCACAGCCCAUCGGUCCGCUCA -------.((..((((((.((((((.(((............((((((..-....))))))))).))).)))...((((...((.((((.....)))).))))))...))))))..)) ( -39.20, z-score = -1.03, R) >droPer1.super_20 890694 109 - 1872136 -------AUGGUCGGAUCACAGUCCUAGACCUUGACUUUGUAGCUCAGC-UUCAUGAGCUUCUUGGAGCUGAUCGGGCAAUUGGGCCACAGGUUGGCACAGCCCAUCGGUCCGCUCA -------.((..((((((.((((((.(((............((((((..-....))))))))).))).)))...((((...((.((((.....)))).))))))...))))))..)) ( -39.20, z-score = -1.03, R) >droWil1.scaffold_180949 5446580 109 + 6375548 ------GGGUUUAAGUUUUA-GAUUUACACACGCACUUUAUAGCUCAAU-UUCAUUAGUUUCUUUGAGCUAAUUGGACAAUUUGGCCAGAGAUUCGCACAACCCAUUGGUCCGCUUA ------(((((((((((...-)))))).....((......((((((((.-.............)))))))).((((.(.....).))))......))..)))))............. ( -20.54, z-score = -0.64, R) >droVir3.scaffold_13049 21314329 112 - 25233164 AUGAUCACAUUAGCCGGCGC----CUAAACACGCACCUUGUAGCUAAGC-UUCAUCAGCUUCUUGGAGCUGAUGGGGCAGUUCGGCCAGAGAUUGGCGCAACCCAUGGGACCGCUCA ..((((.(....(((((((.----.......))).............((-(((((((((((....)))))))))))))....))))..).))))((((...((....))..)))).. ( -38.80, z-score = -0.94, R) >droMoj3.scaffold_6680 15720548 115 - 24764193 AUGAUCACAUUGGCUGGCAU-CAACUAAACGCGCACCUUAUAGCUCAGU-UUCAUCAGCUUCUUGGAGCUGAUGGGGCAGUUGGGCCAGAGAUUGGCACAGCCCAUGGGACCGCUCA ..(.((.(((.(((((...(-(((((....(.((........)).).((-..(((((((((....)))))))))..))))))))(((((...))))).))))).))).)).)..... ( -44.90, z-score = -2.52, R) >droGri2.scaffold_15110 2246786 116 + 24565398 AUGAUCACAUUUACAGUCACACGUCUAAACGCGAACCUUGUAGCUGAGC-UUCAUUAGCUUCUUGGAGCUAAUGGGACAACUCGGCCAGAGAUUGGCACAGCCCAUGGGUCCGCUCA ..............................(((.((((....((((.((-(((((((((((....))))))))))).((((((.....))).))))).))))....)))).)))... ( -35.80, z-score = -1.61, R) >anoGam1.chr2L 10631140 88 - 48795086 ----------------------------AGGUGCACCACGUUGGUUAGC-UUGAGCAGCUUUUUCGAGCUGUACGGACAGCUCGGCCACAGAUGGGAGCAUCCGAUCGGCGUCACGA ----------------------------..((((.((..((.(((.(((-(.....))))....((((((((....))))))))))))).(((.((.....)).))))).).))).. ( -31.60, z-score = -1.08, R) >consensus _______AGGUGCGGAUCCA_UGGCUAGACCCUCACUUUGUAGCUGAGC_UUCAUCAGCUUCUUGGAGCUGAUCGGGCAAUUUGGCCAGAGAUUGGCGCAUCCCAUCGGUCCGCUGA ..........................................((((((..(((((((((((....)))))))).)))...))))))........(((....((....))...))).. (-16.52 = -16.35 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:16 2011