| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,092,234 – 18,092,290 |
| Length | 56 |
| Max. P | 0.996734 |

| Location | 18,092,234 – 18,092,290 |
|---|---|
| Length | 56 |
| Sequences | 8 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 77.25 |
| Shannon entropy | 0.44023 |
| G+C content | 0.46268 |
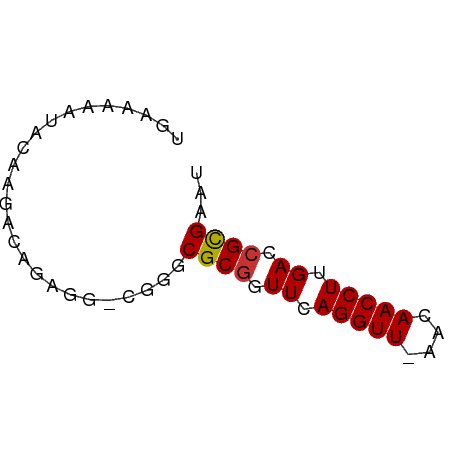
| Mean single sequence MFE | -15.16 |
| Consensus MFE | -10.28 |
| Energy contribution | -10.30 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
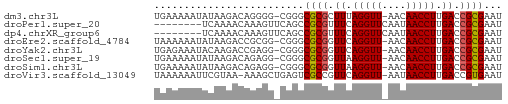
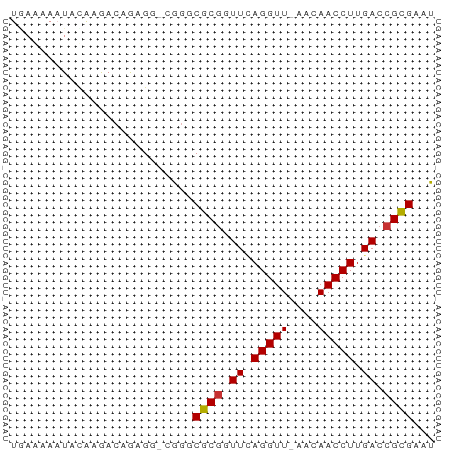
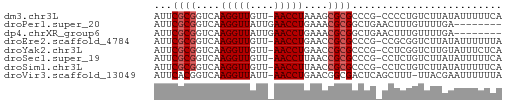
>dm3.chr3L 18092234 56 + 24543557 UGAAAAAUAUAAGACAGGGG-CGGGCGCGCUUUAGGUU-AACAACCUUGACCGCGAAU ................((((-((....)))))).((((-((.....))))))...... ( -10.40, z-score = 0.17, R) >droPer1.super_20 843017 50 + 1872136 --------UCAAAACAAAGUUCAGCCGCGUUUCAGGUUCAAUAACCUUGACCGCGAAU --------.................((((.((.(((((....))))).)).))))... ( -11.00, z-score = -2.34, R) >dp4.chrXR_group6 10932765 50 + 13314419 --------UCAAAACAAAGUUCAGCCGCGUUUCAGGUUCAAUAACCUUGACCGCGAAU --------.................((((.((.(((((....))))).)).))))... ( -11.00, z-score = -2.34, R) >droEre2.scaffold_4784 9963387 56 - 25762168 UAAAAAAUAUAAGACCGCGG-CGGGCGCGGUUCAGGUU-AACAACCUUGACCGCGAAU ..............(((...-))).(((((((.((((.-....)))).)))))))... ( -18.70, z-score = -2.27, R) >droYak2.chr3L 8234111 56 - 24197627 UGAGAAAUACAAGACCGAGG-CGGGCGCGGUUCAGGUU-AACAACCUUGACCGCGAAU ..............(((...-))).(((((((.((((.-....)))).)))))))... ( -18.70, z-score = -2.63, R) >droSec1.super_19 88186 56 + 1257711 UGAAAAAUAUAAGACAGAGG-CGGGCGCGGUUAAGGUU-AACAACCUUGACCGCGAAU ....................-....((((((((((((.-....))))))))))))... ( -20.50, z-score = -4.77, R) >droSim1.chr3L 17419051 56 + 22553184 UGAAAAAUAUAAGACAGAGG-CGGGCGCGGUUAAGGUU-AACAACCUUGACCGCGAAU ....................-....((((((((((((.-....))))))))))))... ( -20.50, z-score = -4.77, R) >droVir3.scaffold_13049 17006381 56 + 25233164 UAAAAAAUUCGUAA-AAAGCUGAGUCGCCGUUCAGGUU-AAUAACCUUGACCGUGAAU ..............-.........((((.(((.((((.-....)))).))).)))).. ( -10.50, z-score = -1.97, R) >consensus UGAAAAAUACAAGACAGAGG_CGGGCGCGGUUCAGGUU_AACAACCUUGACCGCGAAU .........................((((.((.(((((....))))).)).))))... (-10.28 = -10.30 + 0.02)
| Location | 18,092,234 – 18,092,290 |
|---|---|
| Length | 56 |
| Sequences | 8 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Shannon entropy | 0.44023 |
| G+C content | 0.46268 |
| Mean single sequence MFE | -13.43 |
| Consensus MFE | -8.64 |
| Energy contribution | -8.89 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
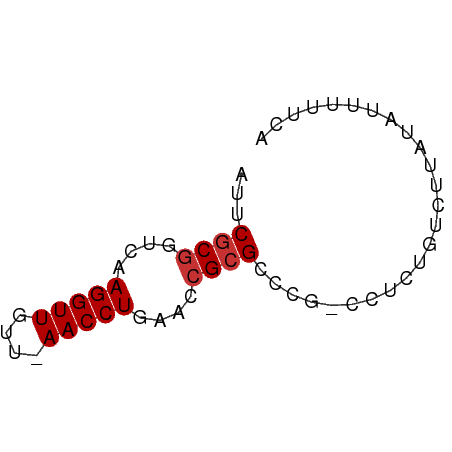
>dm3.chr3L 18092234 56 - 24543557 AUUCGCGGUCAAGGUUGUU-AACCUAAAGCGCGCCCG-CCCCUGUCUUAUAUUUUUCA ...((((.(..((((....-.))))..).))))....-.................... ( -9.70, z-score = -0.29, R) >droPer1.super_20 843017 50 - 1872136 AUUCGCGGUCAAGGUUAUUGAACCUGAAACGCGGCUGAACUUUGUUUUGA-------- ..(((((....(((((....)))))....)))))................-------- ( -12.60, z-score = -1.31, R) >dp4.chrXR_group6 10932765 50 - 13314419 AUUCGCGGUCAAGGUUAUUGAACCUGAAACGCGGCUGAACUUUGUUUUGA-------- ..(((((....(((((....)))))....)))))................-------- ( -12.60, z-score = -1.31, R) >droEre2.scaffold_4784 9963387 56 + 25762168 AUUCGCGGUCAAGGUUGUU-AACCUGAACCGCGCCCG-CCGCGGUCUUAUAUUUUUUA ...((((((..((((....-.))))..)))))).(((-...))).............. ( -16.10, z-score = -1.41, R) >droYak2.chr3L 8234111 56 + 24197627 AUUCGCGGUCAAGGUUGUU-AACCUGAACCGCGCCCG-CCUCGGUCUUGUAUUUCUCA ...((((((..((((....-.))))..)))))).(((-...))).............. ( -16.10, z-score = -1.96, R) >droSec1.super_19 88186 56 - 1257711 AUUCGCGGUCAAGGUUGUU-AACCUUAACCGCGCCCG-CCUCUGUCUUAUAUUUUUCA ...((((((.(((((....-.))))).))))))....-.................... ( -15.70, z-score = -3.88, R) >droSim1.chr3L 17419051 56 - 22553184 AUUCGCGGUCAAGGUUGUU-AACCUUAACCGCGCCCG-CCUCUGUCUUAUAUUUUUCA ...((((((.(((((....-.))))).))))))....-.................... ( -15.70, z-score = -3.88, R) >droVir3.scaffold_13049 17006381 56 - 25233164 AUUCACGGUCAAGGUUAUU-AACCUGAACGGCGACUCAGCUUU-UUACGAAUUUUUUA ((((.((.(((.(((....-.)))))).))((......))...-....))))...... ( -8.90, z-score = -0.93, R) >consensus AUUCGCGGUCAAGGUUGUU_AACCUGAACCGCGCCCG_CCUCUGUCUUAUAUUUUUCA ...((((....(((((....)))))....))))......................... ( -8.64 = -8.89 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:14 2011