
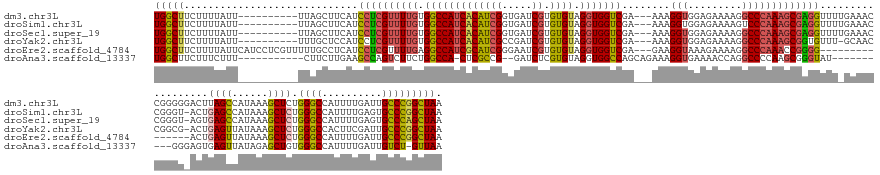
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,085,608 – 18,085,763 |
| Length | 155 |
| Max. P | 0.599460 |

| Location | 18,085,608 – 18,085,763 |
|---|---|
| Length | 155 |
| Sequences | 6 |
| Columns | 168 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Shannon entropy | 0.37732 |
| G+C content | 0.49198 |
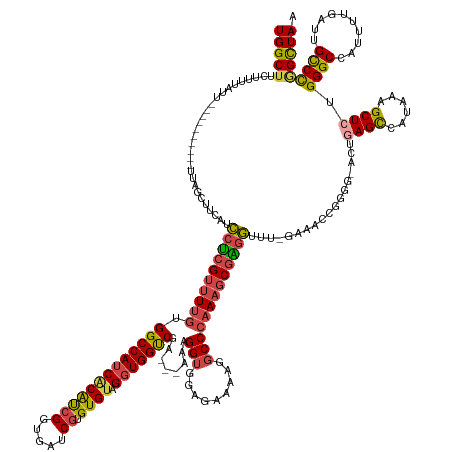
| Mean single sequence MFE | -51.97 |
| Consensus MFE | -33.48 |
| Energy contribution | -33.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18085608 155 + 24543557 UGGCUUCUUUUAUU----------UUAGCUUCAUCCUCGUUUUGUGGCCAUCACAUCGGUGAUCGUGUGUAGGUGGUCGA---AAAGGUGGAGAAAAGGCCCAAAGCGAGGUUUUGAAACCGGGGGACUUAGCCAUAAAGCUCUGGGCCAUUUUGAUUGCCCGGCUAA (((((.......((----------(..(.((((.((((((((((.((((.((.((((..((((((........)))))).---...)))))).....))))))))))))))...)))).)..))).....)))))...((((..((((.((....)).)))))))).. ( -53.00, z-score = -1.30, R) >droSim1.chr3L 17412441 154 + 22553184 UGGCUUCUUUUAUU----------UUAGCUUCAUCCUCGUUUUGUGGCCAUCACAUCGGUGAUCGUGUGUAGGUGGUCGA---AAAGGUGGAGAAAAGUCCCAAAGCGAGGUUUUGAAACCGGGU-ACUGAGCCAUAAAGCUCUGGGCCAUUUUGAGUGCCCGGCUAA ..............----------(((((((((.((((((((((((((((((((((((.....)).)))).)))))))).---......(((......)))))))))))))...))))..(((((-(((((((......)))).....(.....)))))))))))))) ( -53.20, z-score = -2.03, R) >droSec1.super_19 81680 154 + 1257711 UGGCUUCUUUUAUU----------UUAGCUUCAUCCUCGUUUUGUGGCCAUCACAUCGGUGAUCGUGUGUAGGUGGUCGA---AAAGGUGGAGAAAAGGCCCAAAGCGAGGUUUUGAAACCGGGU-AGUGAGCCAUAAAGCUCUGGGCCAUUUUGAGUGCCCAGCUAA ((((((...(((((----------(....((((.((((((((((.((((.((.((((..((((((........)))))).---...)))))).....))))))))))))))...))))...))))-)).))))))...((((..(((((.......).)))))))).. ( -53.80, z-score = -2.14, R) >droYak2.chr3L 8226764 153 - 24197627 UGGCUUCUUUUAUU----------UUUGCUCCAUCCUCGUUUUGUGGCCAUCACAUCGCCGAUCGUGUGUAGGUGGUCGA---AAAGGUGGAGAAAAGGCCCAAAGCGGUGUUU-GCAACCGGCG-ACUGAGUUAUAAAGCUCUGGGCCACUUCGAUUGCCCGGCUAA .(((.((......(----------(((.(((((((....((((.(((((((((((.(((.....)))))).)))))))))---))))))))))))))((((((.((((((....-...)))(((.-.....))).....))).)))))).....))..)))....... ( -51.10, z-score = -0.92, R) >droEre2.scaffold_4784 9956912 150 - 25762168 UGGCUUCUUUUAUUCAUCCUCGUUUUUGCCUCAUCCUCGUUUUGAGGCCAUCGCAUCGGGAAUCGUGUGUAGGUGGUCGA---GAAGGUAAAGAAAAGGCCCAAACCGGGG---------------ACUGAGUUAUAAAGCUCUGGGCCAUUUUGAUUGCCCGGCUAA .(((((.....((((((((((.(((((((((..((.((.....))(((((((((((((.....)).)))).)))))))))---..)))))))))...((......))))))---------------).)))))....)))))((((((.((....)).)))))).... ( -47.70, z-score = -0.61, R) >droAna3.scaffold_13337 18146277 143 - 23293914 UGGCUUCUUUCUUU-----------CUUCUUGAAGCCAGUCUUCUGGCCA-CUCGCCG--GAUCUCGUGUAGGUGGCCAGCAGAAAGGUGAAAACCAGGCCCCAAGCGGGUAU----------GGGAGUGAGUUAUAGAGCUGUGGGCCAUUUUGAUUGUCU-GUUAA (((((((.......-----------......))))))).(((.(((((((-((..(((--.....)).)..))))))))).)))..(((....))).(((((..(((..(((.----------..........)))...)))..))))).............-..... ( -53.02, z-score = -1.69, R) >consensus UGGCUUCUUUUAUU__________UUAGCUUCAUCCUCGUUUUGUGGCCAUCACAUCGGUGAUCGUGUGUAGGUGGUCGA___AAAGGUGGAGAAAAGGCCCAAAGCGAGGUUU_GAAACCGGGG_ACUGAGCCAUAAAGCUCUGGGCCAUUUUGAUUGCCCGGCUAA (((((.............................((((((((((.(((((((((((((.....)).)))).)))))))........(((.........)))))))))))))..................((((......)))).((((..........))))))))). (-33.48 = -33.68 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:10 2011