| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,056,637 – 18,056,760 |
| Length | 123 |
| Max. P | 0.986091 |

| Location | 18,056,637 – 18,056,760 |
|---|---|
| Length | 123 |
| Sequences | 4 |
| Columns | 143 |
| Reading direction | forward |
| Mean pairwise identity | 61.19 |
| Shannon entropy | 0.57566 |
| G+C content | 0.59458 |
| Mean single sequence MFE | -42.77 |
| Consensus MFE | -16.66 |
| Energy contribution | -16.22 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.633854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
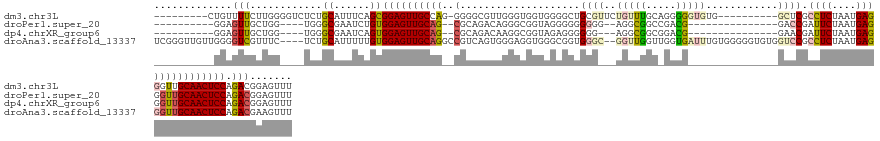
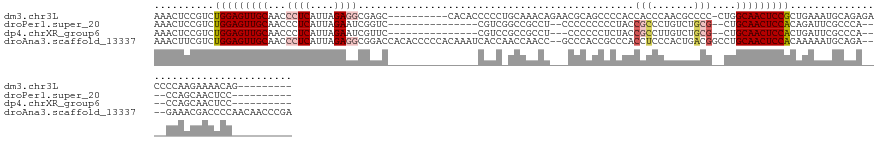
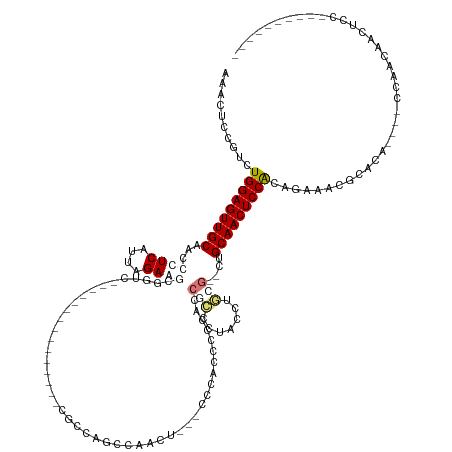
>dm3.chr3L 18056637 123 + 24543557 ---------CUGUUUUCUUGGGGUCUCUGCAUUUCAGCGGAGUUGCCAG-GGGGCGUUGGGUGGUGGGGCUGCGUUCUGUUUGCAGGGGGUGUG----------GCUCGCCUCUAAUGAGGGUUGCAACUCCAGACGGAGUUU ---------....(..(((((.(.((((((......)))))).).))))-)..)(((((((.((((((.((((.(((((....))))).))).)----------.)))))))))))))........((((((....)))))). ( -51.00, z-score = -2.12, R) >droPer1.super_20 807794 110 + 1872136 ----------GGAGUUGCUGG----UGGGCGAAUCUGUGGAGUUGCAG--CGCAGACAGGGCGGUAGGGGGGGGG--AGGCGGCCGACG---------------GACCGAUUCUAAUGAGGGUUGCAACUCCAGACGGAGUUU ----------....(((((..----..))))).(((((((((((((((--(.(....((((((((..(..((...--......))..).---------------.)))).)))).....).)))))))))))..))))).... ( -37.70, z-score = -0.85, R) >dp4.chrXR_group6 10898453 109 + 13314419 ----------GGAGUUGCUGG----UGGGCGAAUCAGUGGAGUUGCAG--CGCAGACAAGGCGGUAGAGGGGGG---AGGCGGCGGACG---------------GAACGAUUCUAAUGAGGGUUGCAACUCCAGACGGAGUUU ----------...((((((((----(......))))))((((((((((--(.(.......((.((.........---..)).))....(---------------((.....))).....).))))))))))).)))....... ( -32.30, z-score = -1.11, R) >droAna3.scaffold_13337 18124994 137 - 23293914 UCGGGUUGUUGGGGUCGUUUC----UCUGCAUUUUUGUGGAGUUGCAGGCCGUCAGUGGGAGGUGGGCGGUGGGC--GGUUGGUUGGUGAUUUGUGGGGGUGUGGUCCGCCUCUAAUGAGGGUUGCAACUCCAGACGAAGUUU (((...(((.((((.....))----)).)))....(.(((((((((((.((.(((...((((((((((..(.(((--.....))).)..(((......)))...))))))))))..))).))))))))))))).))))..... ( -50.10, z-score = -2.26, R) >consensus __________GGAGUUGCUGG____UCGGCAAAUCAGUGGAGUUGCAG__CGCAGACAGGGCGGUAGGGGGGGG___AGGCGGCAGACG_______________GACCGACUCUAAUGAGGGUUGCAACUCCAGACGGAGUUU .............(((............((......))((((((((((..(((..........................)))...........................((((....)))).)))))))))).)))....... (-16.66 = -16.22 + -0.44)
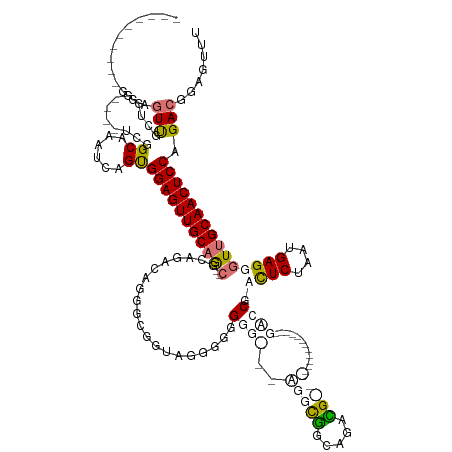
| Location | 18,056,637 – 18,056,760 |
|---|---|
| Length | 123 |
| Sequences | 4 |
| Columns | 143 |
| Reading direction | reverse |
| Mean pairwise identity | 61.19 |
| Shannon entropy | 0.57566 |
| G+C content | 0.59458 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -13.24 |
| Energy contribution | -14.80 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.986091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18056637 123 - 24543557 AAACUCCGUCUGGAGUUGCAACCCUCAUUAGAGGCGAGC----------CACACCCCCUGCAAACAGAACGCAGCCCCACCACCCAACGCCCC-CUGGCAACUCCGCUGAAAUGCAGAGACCCCAAGAAAACAG--------- .......((((((((((((..(((((....)))).)...----------........((((.........))))...................-...))))))))((......))..)))).............--------- ( -29.20, z-score = -2.38, R) >droPer1.super_20 807794 110 - 1872136 AAACUCCGUCUGGAGUUGCAACCCUCAUUAGAAUCGGUC---------------CGUCGGCCGCCU--CCCCCCCCCUACCGCCCUGUCUGCG--CUGCAACUCCACAGAUUCGCCCA----CCAGCAACUCC---------- .......(((((((((((((..........((..(((((---------------....)))))..)--)...........(((.......)))--.)))))))))..))))..((...----...))......---------- ( -25.80, z-score = -1.86, R) >dp4.chrXR_group6 10898453 109 - 13314419 AAACUCCGUCUGGAGUUGCAACCCUCAUUAGAAUCGUUC---------------CGUCCGCCGCCU---CCCCCCUCUACCGCCUUGUCUGCG--CUGCAACUCCACUGAUUCGCCCA----CCAGCAACUCC---------- .......(((((((((((((........((((.......---------------.((.....))..---......)))).(((.......)))--.))))))))))..)))..((...----...))......---------- ( -18.49, z-score = -1.07, R) >droAna3.scaffold_13337 18124994 137 + 23293914 AAACUUCGUCUGGAGUUGCAACCCUCAUUAGAGGCGGACCACACCCCCACAAAUCACCAACCAACC--GCCCACCGCCCACCUCCCACUGACGGCCUGCAACUCCACAAAAAUGCAGA----GAAACGACCCCAACAACCCGA .......(((((((((((((..(((((...((((.((.............................--.........)).))))....))).))..))))))))))((....))....----.....)))............. ( -27.06, z-score = -2.30, R) >consensus AAACUCCGUCUGGAGUUGCAACCCUCAUUAGAAGCGGUC_______________CGCCAGCCAACU___CCCACCCCCACCGCCCUACCUGCG__CUGCAACUCCACAGAAACGCACA____CCAACAACUCC__________ ..........(((((((((...((((....))))..............................................(((.......)))....)))))))))..................................... (-13.24 = -14.80 + 1.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:06 2011