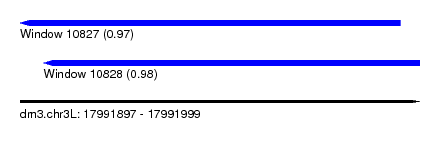
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,991,897 – 17,991,999 |
| Length | 102 |
| Max. P | 0.975439 |

| Location | 17,991,897 – 17,991,994 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 86.80 |
| Shannon entropy | 0.25691 |
| G+C content | 0.56472 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -27.95 |
| Energy contribution | -29.15 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17991897 97 - 24543557 GAAUUCAGGAGCCUGGUAGCACCUGUCACAGAUCUGGAUACUGUGCCACCCACAGGUGUGACCAUGGCGAGUUCUUCGAAUUCUCCCAACUCCUCCG ......(((((((((((.(((((((((((((((....)).)))))......)))))))).)))).)))((((((...)))))).......))))... ( -35.20, z-score = -2.90, R) >droSim1.chr3L 17321554 97 - 22553184 GAAUUCAGGAGCCUGGUAGCACCUGUCACAGAUCUGGAUACUGUGCCACCCACAGGUGUGACCAUGGCGAGUUCUUCGAAUUCUCCCAACUCCUCCG ......(((((((((((.(((((((((((((((....)).)))))......)))))))).)))).)))((((((...)))))).......))))... ( -35.20, z-score = -2.90, R) >droSec1.super_89 94690 97 - 98649 GAAUUCAGGAGCCUGGUAGCACCUGUCACAGAUCUGGAUACUGUGCCACCCACAGGUGUGACCAUGGCGAGUUCUUCGAAUUCUUCCAACUCCUCCG .......((((((((((.(((((((((((((((....)).)))))......)))))))).)))).)))((((((...)))))).))).......... ( -35.20, z-score = -2.95, R) >droYak2.chr3L 8139500 97 + 24197627 GAAUUCAGGAGCCUGGUAGCACCUGUCACAGAUCUGGAUACUGUGCCACCCACAGGUGUGACCAUGGCCAGUUCUUCGAAUUCUCCCAACUCCUCCG (((((((((((((((((.(((((((((((((((....)).)))))......)))))))).)))).)))....)))).)))))).............. ( -34.50, z-score = -2.93, R) >droEre2.scaffold_4784 9873372 97 + 25762168 GAAUUCAGGAGCCUGGUAGCACCUGUCACAGAUCUGGAUACUGUGCCACCCACAGGUGUGACCAUGGCGAGUUCUUCGAAUUCUCCCAACUCCUCCG ......(((((((((((.(((((((((((((((....)).)))))......)))))))).)))).)))((((((...)))))).......))))... ( -35.20, z-score = -2.90, R) >droPer1.super_20 756027 84 - 1872136 ---GCCCGGAGCCUGGUCGCACCUGUCACAGACAUGGAUACUGUGCCGCCCACA-GUGUGCCCAUGGGCAGCAACAGCAGCGGCGGCA--------- ---((((((...)))(((((.((((((...)))).))((((((((.....))))-))))(((....))).((....)).)))))))).--------- ( -32.20, z-score = 0.77, R) >consensus GAAUUCAGGAGCCUGGUAGCACCUGUCACAGAUCUGGAUACUGUGCCACCCACAGGUGUGACCAUGGCGAGUUCUUCGAAUUCUCCCAACUCCUCCG .......((((((((((.(((((((((((((((....)).)))))......)))))))).)))).)))((((((...))))))..........))). (-27.95 = -29.15 + 1.20)



| Location | 17,991,903 – 17,991,999 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 79.02 |
| Shannon entropy | 0.42105 |
| G+C content | 0.55749 |
| Mean single sequence MFE | -33.53 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.57 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17991903 96 - 24543557 AUCCGGAAUU---CAGGAGCCUGGUAGCACCUGUCACAGAUCUGGAUACUGUGCCACCCACAGGUGUGACCAUGGCGAGUUCUUCGAAUUCUCCCAACU ....((((((---((((((((((((.(((((((((((((((....)).)))))......)))))))).)))).)))....)))).)))))))....... ( -35.30, z-score = -2.51, R) >droSim1.chr3L 17321560 96 - 22553184 AUCCGGAAUU---CAGGAGCCUGGUAGCACCUGUCACAGAUCUGGAUACUGUGCCACCCACAGGUGUGACCAUGGCGAGUUCUUCGAAUUCUCCCAACU ....((((((---((((((((((((.(((((((((((((((....)).)))))......)))))))).)))).)))....)))).)))))))....... ( -35.30, z-score = -2.51, R) >droSec1.super_89 94696 96 - 98649 AUCCGGAAUU---CAGGAGCCUGGUAGCACCUGUCACAGAUCUGGAUACUGUGCCACCCACAGGUGUGACCAUGGCGAGUUCUUCGAAUUCUUCCAACU ....((((..---.....(((((((.(((((((((((((((....)).)))))......)))))))).)))).)))((((((...)))))))))).... ( -35.90, z-score = -2.79, R) >droYak2.chr3L 8139506 96 + 24197627 GUCCGGAAUU---CAGGAGCCUGGUAGCACCUGUCACAGAUCUGGAUACUGUGCCACCCACAGGUGUGACCAUGGCCAGUUCUUCGAAUUCUCCCAACU ....((((((---((((((((((((.(((((((((((((((....)).)))))......)))))))).)))).)))....)))).)))))))....... ( -35.30, z-score = -2.42, R) >droEre2.scaffold_4784 9873378 96 + 25762168 AUCCGGAAUU---CAGGAGCCUGGUAGCACCUGUCACAGAUCUGGAUACUGUGCCACCCACAGGUGUGACCAUGGCGAGUUCUUCGAAUUCUCCCAACU ....((((((---((((((((((((.(((((((((((((((....)).)))))......)))))))).)))).)))....)))).)))))))....... ( -35.30, z-score = -2.51, R) >droPer1.super_20 756033 83 - 1872136 ------AUCUGGCCCGGAGCCUGGUCGCACCUGUCACAGACAUGGAUACUGUGCCGCCCACA-GUGUGCCCAUGGGCAGCAACAGCAGCG--------- ------....((((.(....).))))((.((((((...)))).))((((((((.....))))-))))(((....))).((....)).)).--------- ( -27.70, z-score = 1.04, R) >droVir3.scaffold_13049 4055952 79 + 25233164 AGACGGAAUACCAUCAGCGCCAGGUCGCACCUGUCACAGAAUUGGAUACUGUGCCGCCCGCAGGUGUGCCCAUGGCCAG-------------------- .((.((....)).)).(.((((((.((((((((((((((.........)))))......))))))))).)).)))))..-------------------- ( -29.90, z-score = -1.73, R) >consensus AUCCGGAAUU___CAGGAGCCUGGUAGCACCUGUCACAGAUCUGGAUACUGUGCCACCCACAGGUGUGACCAUGGCGAGUUCUUCGAAUUCUCCCAACU ..................(((((((.(((((((((((((.........)))))......)))))))).)))).)))....................... (-23.98 = -24.57 + 0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:59 2011