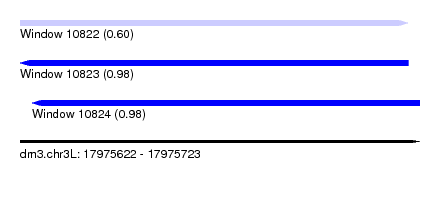
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,975,622 – 17,975,723 |
| Length | 101 |
| Max. P | 0.981301 |

| Location | 17,975,622 – 17,975,720 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 54.79 |
| Shannon entropy | 0.68834 |
| G+C content | 0.36245 |
| Mean single sequence MFE | -19.75 |
| Consensus MFE | -6.07 |
| Energy contribution | -6.82 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
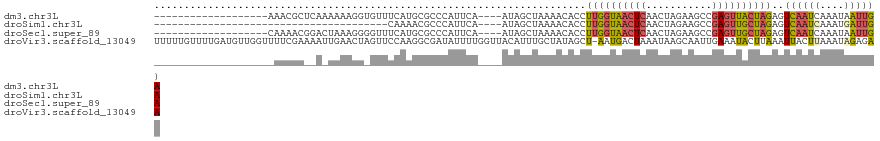
>dm3.chr3L 17975622 98 + 24543557 -------------------AAACGCUCAAAAAAGGUGUUUCAUGCGCCCAUUCA----AUAGCUAAAACACCUUGGUAACUCAACUAGAAGCCGAGUUACUAGAGUCAAUCAAAUAAUUGA -------------------....((((....(((((((((...((.........----...))..)))))))))((((((((...........)))))))).))))((((......)))). ( -24.50, z-score = -3.50, R) >droSim1.chr3L 17300875 78 + 22553184 ---------------------------------------CAAAACGCCCAUUCA----AUAGCUAAAACACCUUGGUAACUCAACUAGAAGCCGAGUUGCUAGAGUCAAUCAAAUGAUUGA ---------------------------------------...............----..............((((((((((...........))))))))))..((((((....)))))) ( -14.90, z-score = -1.74, R) >droSec1.super_89 78654 98 + 98649 -------------------CAAAACGGACUAAAGGGGUUUCAUGCGCCCAUUCA----AUAGCUAAAACACCUUGGUAACUCAACUAGAAGCCGAGUUGCUAGAGUCAAUCAAAUAAUUGA -------------------.......((((...(((((.....)).))).....----..............((((((((((...........)))))))))))))).............. ( -20.20, z-score = -0.74, R) >droVir3.scaffold_13049 4038633 120 - 25233164 UUUUUGUUUUGAUGUUGGUUUUCGAAAAUUGAACUAGUUCCAAGGCGAUAUUUUGGUUACAUUUGCUAUAGCU-AAUGACUAAAUAAGCAAUUGAAAUACUUAAAUUACUUAAAUAGAGAA (((((((((((((...(((((((((.....(((....)))....((.....((((((..((((.((....)).-))))))))))...))..)))))).)))...))))...))))))))). ( -19.40, z-score = -0.23, R) >consensus ____________________AACGC__A_UAAAGG_GUUCCAAGCGCCCAUUCA____AUAGCUAAAACACCUUGGUAACUCAACUAGAAGCCGAGUUACUAGAGUCAAUCAAAUAAUUGA ........................................................................((((((((((...........))))))))))..((((((....)))))) ( -6.07 = -6.82 + 0.75)
| Location | 17,975,622 – 17,975,720 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 54.79 |
| Shannon entropy | 0.68834 |
| G+C content | 0.36245 |
| Mean single sequence MFE | -21.55 |
| Consensus MFE | -9.68 |
| Energy contribution | -8.43 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
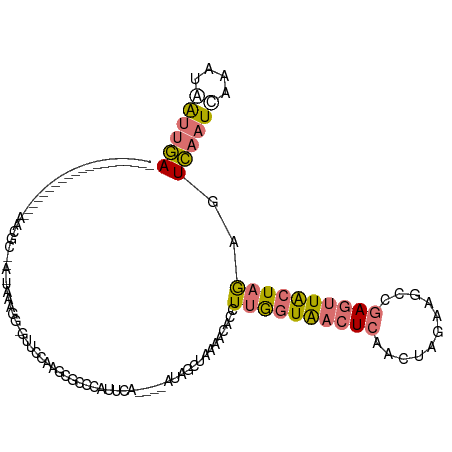
>dm3.chr3L 17975622 98 - 24543557 UCAAUUAUUUGAUUGACUCUAGUAACUCGGCUUCUAGUUGAGUUACCAAGGUGUUUUAGCUAU----UGAAUGGGCGCAUGAAACACCUUUUUUGAGCGUUU------------------- ((((((....))))))(((..((((((((((.....)))))))))).((((((((((((((..----......)))...)))))))))))....))).....------------------- ( -30.90, z-score = -3.74, R) >droSim1.chr3L 17300875 78 - 22553184 UCAAUCAUUUGAUUGACUCUAGCAACUCGGCUUCUAGUUGAGUUACCAAGGUGUUUUAGCUAU----UGAAUGGGCGUUUUG--------------------------------------- ((((((....)))))).......((((((((.....))))))))..(((..(((((...(...----.)...)))))..)))--------------------------------------- ( -18.50, z-score = -1.17, R) >droSec1.super_89 78654 98 - 98649 UCAAUUAUUUGAUUGACUCUAGCAACUCGGCUUCUAGUUGAGUUACCAAGGUGUUUUAGCUAU----UGAAUGGGCGCAUGAAACCCCUUUAGUCCGUUUUG------------------- ((((((....))))))...((((((((((((.....))))))))((....))......)))).----.((((((((....(......)....))))))))..------------------- ( -23.20, z-score = -1.14, R) >droVir3.scaffold_13049 4038633 120 + 25233164 UUCUCUAUUUAAGUAAUUUAAGUAUUUCAAUUGCUUAUUUAGUCAUU-AGCUAUAGCAAAUGUAACCAAAAUAUCGCCUUGGAACUAGUUCAAUUUUCGAAAACCAACAUCAAAACAAAAA ....(((..(((((((((..........)))))))))..))).((((-.((....)).))))...........(((.....(((....)))......)))..................... ( -13.60, z-score = -0.65, R) >consensus UCAAUUAUUUGAUUGACUCUAGCAACUCGGCUUCUAGUUGAGUUACCAAGGUGUUUUAGCUAU____UGAAUGGGCGCAUGAAAC_CCUUUA_U__GCGUU____________________ ((((((....)))))).....((((((((((.....)))))))))).............((((.......))))............................................... ( -9.68 = -8.43 + -1.25)
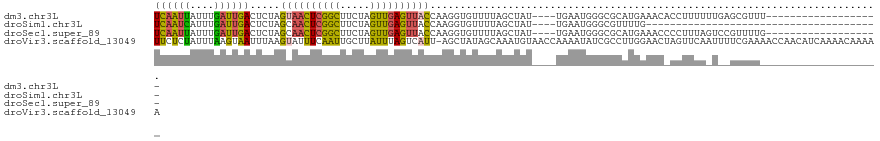
| Location | 17,975,625 – 17,975,723 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 58.96 |
| Shannon entropy | 0.67610 |
| G+C content | 0.36585 |
| Mean single sequence MFE | -20.18 |
| Consensus MFE | -9.08 |
| Energy contribution | -9.75 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
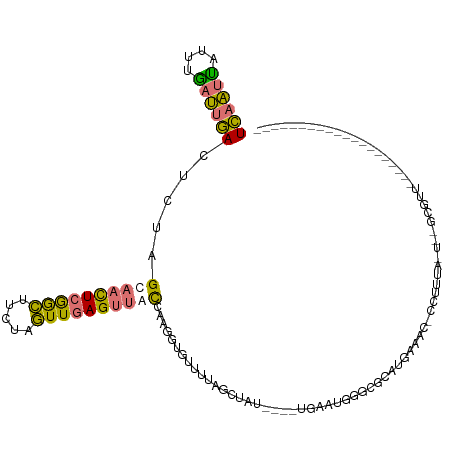
>dm3.chr3L 17975625 98 - 24543557 ACUUCAAUUAUUUGAUUGACUCUAGUAACUCGGCUUCUAGUUGAGUUACCA-----AGGUGUUUUAGCUAUUGAAUGGGCGCAUGAAACACCUUUUUUGAGCG----------------- ...((((((....))))))(((..((((((((((.....)))))))))).(-----(((((((((((((........)))...)))))))))))....)))..----------------- ( -31.10, z-score = -4.01, R) >droSim1.chr3L 17300878 78 - 22553184 ACUUCAAUCAUUUGAUUGACUCUAGCAACUCGGCUUCUAGUUGAGUUACCA-----AGGUGUUUUAGCUAUUGAAUGGGCGUU------------------------------------- ...((((((....))))))(..((((((((((((.....))))))))((..-----..))......))))..)..........------------------------------------- ( -18.60, z-score = -1.12, R) >droSec1.super_89 78657 98 - 98649 ACUUCAAUUAUUUGAUUGACUCUAGCAACUCGGCUUCUAGUUGAGUUACCA-----AGGUGUUUUAGCUAUUGAAUGGGCGCAUGAAACCCCUUUAGUCCGUU----------------- ...((((((....))))))(..((((((((((((.....))))))))((..-----..))......))))..)(((((((....(......)....)))))))----------------- ( -22.70, z-score = -0.88, R) >droYak2.chr3L 8123108 75 + 24197627 ACUCCAAUUAUUUGAUUGACUCUAUUAACUCGGCUUCCAGUUGAGUUAUUA-----AGGUGUUUUCACUAUUGAAUGGGU---------------------------------------- ..((((.(((..(((..(((..((.(((((((((.....))))))))).))-----..)).)..)))....))).)))).---------------------------------------- ( -17.70, z-score = -1.99, R) >droEre2.scaffold_4784 9857183 76 + 25762168 ACUCCAACUAUUUGAUUGACUCUAUUAACUCGGCUUCCAGUUGAGUUAUCA-----AGGUGUUUUAGCUUUUGAAUGGGUC--------------------------------------- .................(((((...(((((((((.....)))))))))(((-----(((.((....))))))))..)))))--------------------------------------- ( -17.40, z-score = -1.39, R) >droVir3.scaffold_13049 4038636 117 + 25233164 ---UUCUCUAUUUAAGUAAUUUAAGUAUUUCAAUUGCUUAUUUAGUCAUUAGCUAUAGCAAAUGUAACCAAAAUAUCGCCUUGGAACUAGUUCAAUUUUCGAAAACCAACAUCAAAACAA ---....(((..(((((((((..........)))))))))..))).((((.((....)).))))...........(((.....(((....)))......))).................. ( -13.60, z-score = -0.67, R) >consensus ACUUCAAUUAUUUGAUUGACUCUAGUAACUCGGCUUCUAGUUGAGUUACCA_____AGGUGUUUUAGCUAUUGAAUGGGCG_______________________________________ .......((((((((.........((((((((((.....))))))))))........(((......))).)))))))).......................................... ( -9.08 = -9.75 + 0.67)

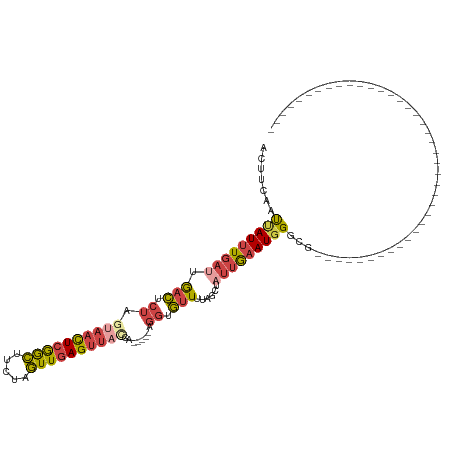
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:56 2011