| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,612,084 – 5,612,185 |
| Length | 101 |
| Max. P | 0.803068 |

| Location | 5,612,084 – 5,612,185 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.76 |
| Shannon entropy | 0.47886 |
| G+C content | 0.42070 |
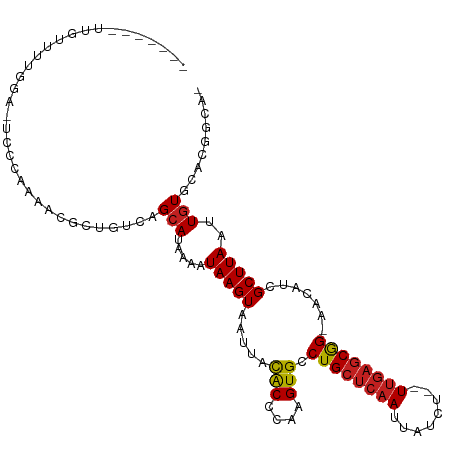
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -11.91 |
| Energy contribution | -11.89 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5612084 101 + 23011544 -------UUGUUUUGGG-UCCCAGGGCGCUGUCAGCAUAAAGUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCU--UUGAGCGG-AACAUUGCUUAAUUGUGCACGGCA- -------..(((((((.-..)))))))(((((..((((((..((((((((..(((....))).((((((((......--))))))))-...)))))))).))))))))))).- ( -36.20, z-score = -2.95, R) >droPer1.super_10 1194670 103 - 3432795 -------UCGUUUCCCAACACACACACGAUGUCAGCAUAAAAUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCU--UUGAGCGGGAACAUCGCUUAAUUGUCCAUGGCA- -------((((..............))))(((((.....((.(((((.....(((....)))(((((((((......--)))))))))......))))).)).....)))))- ( -23.84, z-score = -2.01, R) >dp4.chr4_group4 3370097 103 - 6586962 -------UUGUUUCCCAACACACACACGAUGUCAGCAUAAAAUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCU--UUGAGCGGGAACAUCGCUUAAUUGUCCAUGGCA- -------.......((((((......((((((....................(((....)))(((((((((......--))))))))).))))))......)))...)))..- ( -23.60, z-score = -1.70, R) >droAna3.scaffold_12916 11057799 102 + 16180835 --------UGUUUUGGGGUCCCAGCAGGCUGUCAGCAUAAAGUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCU--UUGAGCGG-AACAUCGCUUAAUUGUGCACGGCAG --------(((..(((....)))))).(((((..((((((.((((....))))....(((((.((((((((......--))))))))-.....)))))..))))))))))).. ( -32.90, z-score = -2.07, R) >droEre2.scaffold_4929 5700537 101 + 26641161 -------UUGUUUUGGG-UCCCCAAACGCUGUCAGCAUAAAGUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCU--UUGAGCGG-AACAUUGCUUAAUUGUGCACGGCA- -------....(((((.-...))))).(((((..((((((..((((((((..(((....))).((((((((......--))))))))-...)))))))).))))))))))).- ( -35.40, z-score = -3.85, R) >droYak2.chr2L 8741979 101 - 22324452 -------UUGUUUUGGG-UCCCAGAGCCCUGUCAGCAUAAAGUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCU--UUGAGCGG-AACAUUGCUUAAUUGUGCACGGCA- -------..(((((((.-..)))))))...(((.((((((..((((((((..(((....))).((((((((......--))))))))-...)))))))).))))))..))).- ( -31.90, z-score = -2.49, R) >droSec1.super_5 3689634 101 + 5866729 -------UUGGUUUGGG-UCCCAGAGCGCUGUCAGCAUAAAGUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCU--UUGAGCGG-AACAUUGCUUAAUUGUGCACGGCA- -------..(.(((((.-..))))).)(((((..((((((..((((((((..(((....))).((((((((......--))))))))-...)))))))).))))))))))).- ( -35.00, z-score = -2.73, R) >droSim1.chr2L 5406377 101 + 22036055 -------UUGUUUUGGG-UCCCAGAGCGCUGUCAGCAUAAAGUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCU--UUGAGCGG-AACAUUGCUUAAUUGUGCACGGCA- -------..(((((((.-..)))))))(((((..((((((..((((((((..(((....))).((((((((......--))))))))-...)))))))).))))))))))).- ( -37.10, z-score = -3.78, R) >droWil1.scaffold_180708 3896521 108 - 12563649 ----UUGAUAUGGAAAAUCCCAUUACAGGUGUCAGAAUAAAAUAAGUAAUUAUGCCCAAGUGCCUGCUCAAUUAUCUCAUCGUACAG-AAAAUCGCUUAAUUGUACAGGGUAG ----..............(((....((((..(.........((((....))))......)..))))...............((((((-............)))))).)))... ( -15.56, z-score = 0.82, R) >droVir3.scaffold_12963 3573155 98 + 20206255 UUUGGGUCUA--GGUAA-ACCGAAAAUACUGUCAGCAUAAAAUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCC--UUGAGCAG-CAUAACGCUUAAUUGU--------- .((((((.((--((...-.)).....((((..............))))..)).))))))((((.(((((((......--))))))))-))).............--------- ( -23.64, z-score = -2.21, R) >droMoj3.scaffold_6500 13966123 103 + 32352404 UUUGGUUCUG--GAAAAUAACCGAAAUGCUGUCAGCAUAUAAUAAGUAAUUAUACCCAAGUGCCUGCUCAAUUAUCU--UUAAGCAG-CAUAACGCUUAAUUGUGCAU----- (((((((.(.--....).))))))).........(((((..((((....))))......((((.((((.((......--)).)))))-)))..........)))))..----- ( -18.90, z-score = -0.09, R) >droGri2.scaffold_15252 3110185 108 + 17193109 -UCGGCACUGCCAGAAAAACUGAACAUACUGUCAGCAUAAAAUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCU--UUGAGCAG-AACAUCGCUUAAUUGUGCAAAGCG- -..((((.((.(((.....)))..))...)))).((((((..(((((.....(((....))).((((((((......--))))))))-......))))).))))))......- ( -25.80, z-score = -2.15, R) >consensus _______UUGUUUUGGA_UCCCAAAACGCUGUCAGCAUAAAAUAAGUAAUUACACCCAAGUGCCUGCUCAAUUAUCU__UUGAGCGG_AACAUCGCUUAAUUGUGCACGGCA_ ..................................(((.....(((((.....(((....))).((((((((........)))))))).......)))))..)))......... (-11.91 = -11.89 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:29 2011