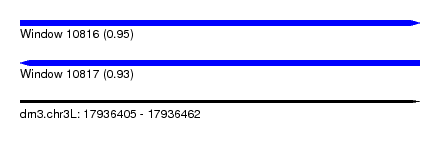
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,936,405 – 17,936,462 |
| Length | 57 |
| Max. P | 0.949969 |

| Location | 17,936,405 – 17,936,462 |
|---|---|
| Length | 57 |
| Sequences | 5 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 74.57 |
| Shannon entropy | 0.44578 |
| G+C content | 0.38011 |
| Mean single sequence MFE | -14.10 |
| Consensus MFE | -7.88 |
| Energy contribution | -8.40 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.949969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
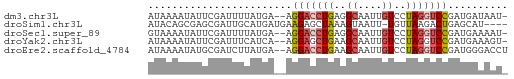
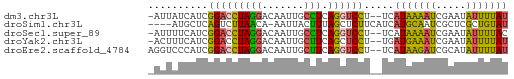
>dm3.chr3L 17936405 57 + 24543557 AUAAAAUAUUCGAUUUUAUGA--AGGACCUGAGGCAAUUGUCCUAGGUCCGAUGAUAAU- (((((((.....)))))))..--.(((((((.(((....))).))))))).........- ( -16.70, z-score = -3.23, R) >droSim1.chr3L 17261247 55 + 22553184 AUACAGCGAGCGAUUGCAUGAUGAAGAGCUAAAGUAAUU-UGUUAAGACUGAGCAU---- .....((((....)))).(((..((..((....))..))-..)))...........---- ( -6.90, z-score = 0.76, R) >droSec1.super_89 40219 57 + 98649 GUAAAAUAUUCGAUUUUAUGA--AGGACCUGAGGCAAUUGUCCUAGGUCCGAUGAAAAU- (((((((.....)))))))..--.(((((((.(((....))).))))))).........- ( -17.00, z-score = -3.44, R) >droYak2.chr3L 8080358 57 - 24197627 AUAAAAUAUUCGAUUUCAUCA--AGGAGCUGAAGCAAUUGUCCUAGGUCCGAUGAAAGU- .............(((((((.--.(((.(((..((....))..))).))))))))))..- ( -13.90, z-score = -2.31, R) >droEre2.scaffold_4784 9817503 58 - 25762168 AUAAAAUAUGCGAUCUUAUGA--AGGACCUGAAGCAAUUGUCCUAGGUCCGAUGGGACCU ...........(.((((((..--.(((((((..((....))..))))))).)))))).). ( -16.00, z-score = -2.16, R) >consensus AUAAAAUAUUCGAUUUUAUGA__AGGACCUGAAGCAAUUGUCCUAGGUCCGAUGAAAAU_ ........................(((((((..((....))..))))))).......... ( -7.88 = -8.40 + 0.52)
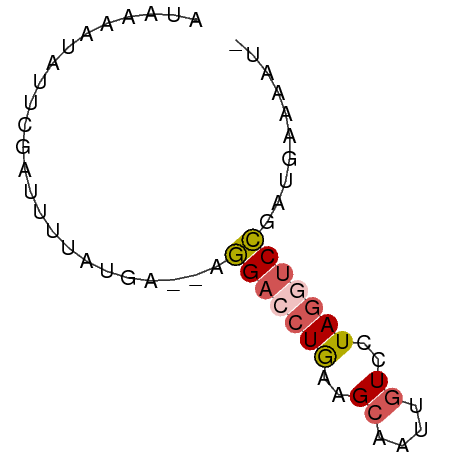
| Location | 17,936,405 – 17,936,462 |
|---|---|
| Length | 57 |
| Sequences | 5 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 74.57 |
| Shannon entropy | 0.44578 |
| G+C content | 0.38011 |
| Mean single sequence MFE | -10.52 |
| Consensus MFE | -8.18 |
| Energy contribution | -7.62 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
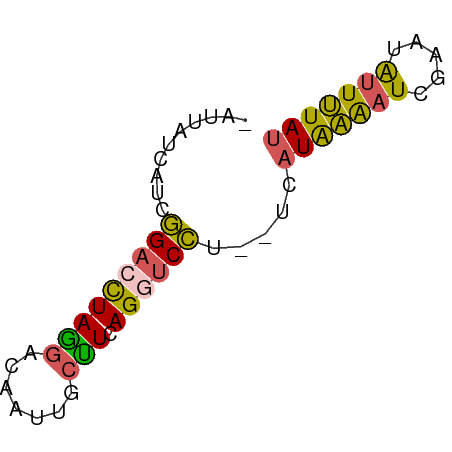
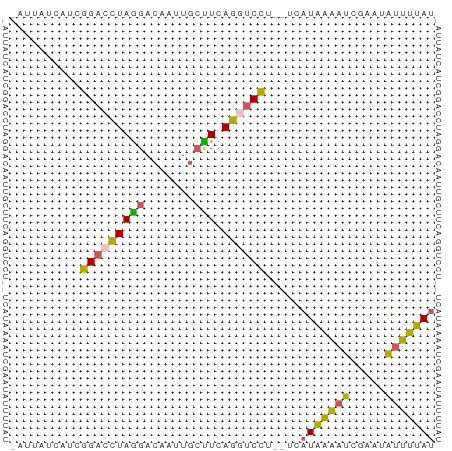
>dm3.chr3L 17936405 57 - 24543557 -AUUAUCAUCGGACCUAGGACAAUUGCCUCAGGUCCU--UCAUAAAAUCGAAUAUUUUAU -.........(((((((((.(....)))).)))))).--..(((((((.....))))))) ( -13.90, z-score = -2.97, R) >droSim1.chr3L 17261247 55 - 22553184 ----AUGCUCAGUCUUAACA-AAUUACUUUAGCUCUUCAUCAUGCAAUCGCUCGCUGUAU ----..(((.(((..(....-.)..)))..)))........(((((.........))))) ( -1.40, z-score = 2.20, R) >droSec1.super_89 40219 57 - 98649 -AUUUUCAUCGGACCUAGGACAAUUGCCUCAGGUCCU--UCAUAAAAUCGAAUAUUUUAC -.........(((((((((.(....)))).)))))).--..................... ( -13.50, z-score = -2.91, R) >droYak2.chr3L 8080358 57 + 24197627 -ACUUUCAUCGGACCUAGGACAAUUGCUUCAGCUCCU--UGAUGAAAUCGAAUAUUUUAU -..((((((((((.(((((.(....)))).)).))).--.)))))))............. ( -9.90, z-score = -1.18, R) >droEre2.scaffold_4784 9817503 58 + 25762168 AGGUCCCAUCGGACCUAGGACAAUUGCUUCAGGUCCU--UCAUAAGAUCGCAUAUUUUAU ((((((....))))))(((((...........)))))--..(((((((.....))))))) ( -13.90, z-score = -1.70, R) >consensus _AUUAUCAUCGGACCUAGGACAAUUGCUUCAGGUCCU__UCAUAAAAUCGAAUAUUUUAU ..........(((((((((.......))).)))))).....(((((((.....))))))) ( -8.18 = -7.62 + -0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:51 2011