
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,892,263 – 17,892,366 |
| Length | 103 |
| Max. P | 0.949681 |

| Location | 17,892,263 – 17,892,362 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.99 |
| Shannon entropy | 0.53813 |
| G+C content | 0.49154 |
| Mean single sequence MFE | -24.16 |
| Consensus MFE | -12.89 |
| Energy contribution | -12.89 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
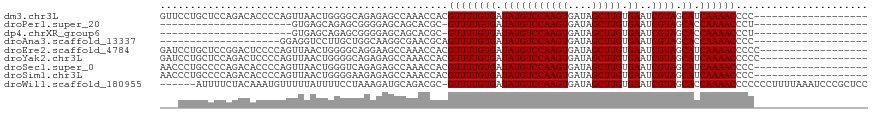
>dm3.chr3L 17892263 99 - 24543557 GUUCCUGCUCCAGACACCCCAGUUAACUGGGGCAGAGAGCCAAACCACGUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCAUCAAAACCCC------------------- (((((((((((((((......))...))))))))).))))........((((((((.(((((((((((....))))).))..)))).)).))))))...------------------- ( -32.10, z-score = -3.69, R) >droPer1.super_20 660910 76 - 1872136 ----------------------GUGAGCAGAGCGGGGAGCAGCACGC-GUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCACCAAAACCCU------------------- ----------------------(((.((((((((.(........).)-)))))))..(((((((((((....))))).))..)))).))).........------------------- ( -18.90, z-score = 0.10, R) >dp4.chrXR_group6 10748387 76 - 13314419 ----------------------GUGAGCAGAGCGGGGAGCAGCACGC-GUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCACCAAAACCCU------------------- ----------------------(((.((((((((.(........).)-)))))))..(((((((((((....))))).))..)))).))).........------------------- ( -18.90, z-score = 0.10, R) >droAna3.scaffold_13337 17986826 79 + 23293914 --------------------GGAGGUCCUUGCUGGCAAGGCGAACGCAGUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCACCAAAACCCC------------------- --------------------((..(.(((((....)))))).......((((((((.(((((((((((....))))).))..)))).)).)))))))).------------------- ( -20.30, z-score = -0.34, R) >droEre2.scaffold_4784 9775170 100 + 25762168 GAUCCUGCUCCGGACUCCCCAGUUAACUGGGGCAGGAAGCCAAACCACGUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCAUCAAAACCCCC------------------ ..((((((((((((((....)))...)))))))))))...........((((((((.(((((((((((....))))).))..)))).)).))))))....------------------ ( -32.40, z-score = -2.92, R) >droYak2.chr3L 8037356 100 + 24197627 GAUCCUGCUCCAGACUCCCCAGUUAACUGGGGCAGAGAGCCAAACCACGUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCAUCAAAACCCCC------------------ ......((((....((((((((....)))))).)).))))........((((((((.(((((((((((....))))).))..)))).)).))))))....------------------ ( -29.50, z-score = -2.95, R) >droSec1.super_0 9921824 99 - 21120651 AACCCUGCCCCAGACACCCCAGUUAACUGGGUCAGAGAGCCAAACCACGUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCAUCAAAACCCC------------------- .....(((..(.((((.(((((....)))))(((.(((((........))))).)))..))))(((((....)))))......)..)))..........------------------- ( -23.70, z-score = -2.22, R) >droSim1.chr3L 17219357 99 - 22553184 AACCCUGCCCCAGACACCCCAGUUAACUGGGGAAGAGAGCCAAACCACGUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCAUCAAAACCCC------------------- ................((((((....))))))................((((((((.(((((((((((....))))).))..)))).)).))))))...------------------- ( -25.00, z-score = -2.23, R) >droWil1.scaffold_180955 235359 111 + 2875958 ------AUUUUCUACAAAUGUUUUUAUUUUCCUAAAGAUGCAGACGC-GUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCACCAAAACCCCCCCUUUUAAAUCCCGCUCC ------.............((((.((((((....)))))).))))..-((((((((.(((((((((((....))))).))..)))).)).))))))...................... ( -16.60, z-score = -1.35, R) >consensus ___CCUGCUCCAGACACCCCAGUUAACUGGGGCAGAGAGCCAAACCACGUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCAUCAAAACCCC___________________ ................................................((((((((.(((((((((((....))))).))..)))).)).))))))...................... (-12.89 = -12.89 + -0.00)

| Location | 17,892,263 – 17,892,366 |
|---|---|
| Length | 103 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.48 |
| Shannon entropy | 0.56372 |
| G+C content | 0.50109 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -13.04 |
| Energy contribution | -12.96 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.948911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
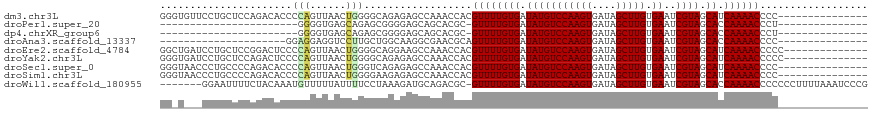
>dm3.chr3L 17892263 103 - 24543557 GGGUGUUCCUGCUCCAGACACCCCAGUUAACUGGGGCAGAGAGCCAAACCACGUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCAUCAAAACCCC--------------- (((.(((((((((((((((......))...))))))))).))))........((((((((.(((((((((((....))))).))..)))).)).)))))))))--------------- ( -35.00, z-score = -3.02, R) >droPer1.super_20 660910 79 - 1872136 -----------------------GGGGUGAGCAGAGCGGGGAGCAGCACGC-GUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCACCAAAACCCU--------------- -----------------------(((((..((((((((.(........).)-)))))))..(((((((((((....))))).))..))))........)))))--------------- ( -23.40, z-score = -0.54, R) >dp4.chrXR_group6 10748387 79 - 13314419 -----------------------GGGGUGAGCAGAGCGGGGAGCAGCACGC-GUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCACCAAAACCCU--------------- -----------------------(((((..((((((((.(........).)-)))))))..(((((((((((....))))).))..))))........)))))--------------- ( -23.40, z-score = -0.54, R) >droAna3.scaffold_13337 17986826 82 + 23293914 ---------------------GGAGGAGGUCCUUGCUGGCAAGGCGAACGCAGUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCACCAAAACCCC--------------- ---------------------...((..(.(((((....)))))).......((((((((.(((((((((((....))))).))..)))).)).)))))))).--------------- ( -20.50, z-score = 0.14, R) >droEre2.scaffold_4784 9775170 104 + 25762168 GGCUGAUCCUGCUCCGGACUCCCCAGUUAACUGGGGCAGGAAGCCAAACCACGUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCAUCAAAACCCCC-------------- ((((..((((((((((((((....)))...))))))))))))))).......((((((((.(((((((((((....))))).))..)))).)).))))))....-------------- ( -38.50, z-score = -3.82, R) >droYak2.chr3L 8037356 104 + 24197627 GGGUGAUCCUGCUCCAGACUCCCCAGUUAACUGGGGCAGAGAGCCAAACCACGUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCAUCAAAACCCCC-------------- (((((.((((((((((((((....)))...))))))))).))..))......((((((((.(((((((((((....))))).))..)))).)).))))))))).-------------- ( -33.60, z-score = -2.62, R) >droSec1.super_0 9921824 103 - 21120651 GGGUAACCCUGCCCCAGACACCCCAGUUAACUGGGUCAGAGAGCCAAACCACGUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCAUCAAAACCCC--------------- ((((.....(((..(.((((.(((((....)))))(((.(((((........))))).)))..))))(((((....)))))......)..))).....)))).--------------- ( -29.20, z-score = -2.44, R) >droSim1.chr3L 17219357 103 - 22553184 GGGUAACCCUGCCCCAGACACCCCAGUUAACUGGGGAAGAGAGCCAAACCACGUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCAUCAAAACCCC--------------- (((((....)))))......((((((....))))))................((((((((.(((((((((((....))))).))..)))).)).))))))...--------------- ( -30.80, z-score = -2.63, R) >droWil1.scaffold_180955 235363 110 + 2875958 -------GGAAUUUUCUACAAAUGUUUUUAUUUUCCUAAAGAUGCAGACGC-GUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCACCAAAACCCCCCCUUUUAAAUCCCG -------(((.............((((.((((((....)))))).))))..-((((((((.(((((((((((....))))).))..)))).)).)))))).............))).. ( -19.10, z-score = -1.66, R) >consensus GG_U___CCUGCUCCAGACACCCCAGUUAACUGGGGCAGAGAGCCAAACCACGUUUUGUGAUAUGUCCAAGUGAUAGCUUGUGAAUCGUAGCAUCAAAACCCC_______________ ......................(((......)))..................((((((((.(((((((((((....))))).))..)))).)).)))))).................. (-13.04 = -12.96 + -0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:46 2011