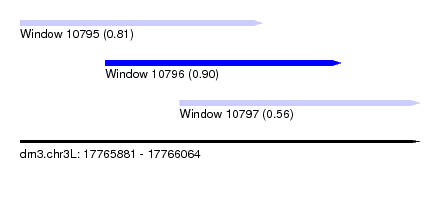
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,765,881 – 17,766,064 |
| Length | 183 |
| Max. P | 0.901663 |

| Location | 17,765,881 – 17,765,992 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 95.59 |
| Shannon entropy | 0.07560 |
| G+C content | 0.45045 |
| Mean single sequence MFE | -30.71 |
| Consensus MFE | -28.04 |
| Energy contribution | -27.80 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.807750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
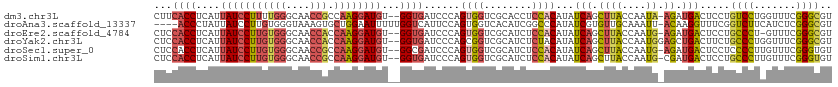
>dm3.chr3L 17765881 111 + 24543557 GAAUGAAAUUUAAACCCGAAACUGAGCGGUAAUUACAUCAUACAUGGCUCGAUAGUUUUCCAAGUGGCGUCAGCUUCACCUCAUUAUCCUUUUGGGCAACCGCCAAGGAUG .(((((...........((((((((((.((.............)).))))...))))))....(.(((....))).)...)))))((((((.(((....)))..)))))). ( -23.72, z-score = -0.15, R) >droEre2.scaffold_4784 9667833 111 - 25762168 GAAUGAAAUUUAAAUCCGAAACUGAGUGGUAAUUACAUCAUGCAAGGCUCGAUAGUUUUCCAAGUGGCGGCAGCUCCACCUCAUUAUCCUUGUGGGCAACCACCAAGGAUG .(((((..........(((..((..(((((......)))))...))..)))............((((.(....).)))).)))))((((((((((....))).))))))). ( -30.80, z-score = -1.50, R) >droYak2.chr3L 7905893 111 - 24197627 GAAUGAAAUUUAAAUCCGAAACUGAGUGGUAAUUACAUCAUUCAUGGCUCGAUAGUUUUCCAAGUGGCGGCAGCUCCACCUCAUUAUCCUUGUGGGCAACCACCAAGGAUG .(((((..........(((..(((((((((......)))))))).)..)))............((((.(....).)))).)))))((((((((((....))).))))))). ( -33.60, z-score = -2.55, R) >droSec1.super_0 9812971 111 + 21120651 GAAUGAAAUUUAAACCCGAAACUGAGCGGUAAUUACAUCAUACAUGGCUCGAUAGUUUUGCAAGUGGCGUCAGCUCCACCUCAUUAUCCUUGUGGGCAACCGCCAAGGAUG .(((((..........(((((((((((.((.............)).))))...)))))))...((((.(....).)))).)))))((((((((((....))).))))))). ( -32.72, z-score = -2.25, R) >droSim1.chr3L 17103530 111 + 22553184 GAAUGAAAUUUAAACCCGAAACUGAGCGGUAAUUACAUCAUACAUGGCUCGAUAGUUUUGCAAGUGGCGUCAGCUCCACCUCAUUAUCCUUGUGGGCAACCGCCAAGGAUG .(((((..........(((((((((((.((.............)).))))...)))))))...((((.(....).)))).)))))((((((((((....))).))))))). ( -32.72, z-score = -2.25, R) >consensus GAAUGAAAUUUAAACCCGAAACUGAGCGGUAAUUACAUCAUACAUGGCUCGAUAGUUUUCCAAGUGGCGUCAGCUCCACCUCAUUAUCCUUGUGGGCAACCGCCAAGGAUG .(((((...........((((((((((.((.............)).))))...))))))....((((.(....).)))).)))))((((((((((....))).))))))). (-28.04 = -27.80 + -0.24)

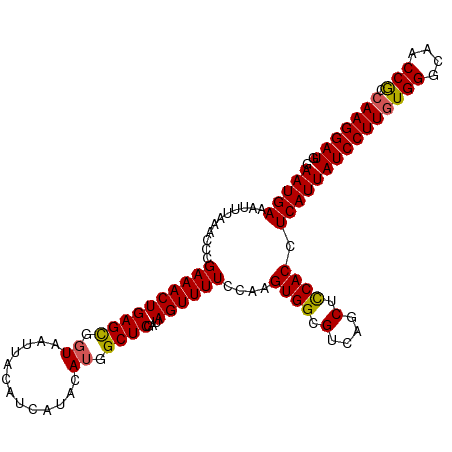
| Location | 17,765,920 – 17,766,028 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 95.09 |
| Shannon entropy | 0.08646 |
| G+C content | 0.52037 |
| Mean single sequence MFE | -36.33 |
| Consensus MFE | -33.12 |
| Energy contribution | -32.76 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17765920 108 + 24543557 AUACAUGGCUCGAUAGUUUUCCAAGUGGCGUCAGCUUCACCUCAUUAUCCUUUUGGGCAACCGCCAAGGAUGUGGUGAUCCCAGUGGUCGCACCUCCACAUAUCAGCU ......((((.((((...........(((....)))(((((....(((((((.(((....)))..))))))).))))).....((((........)))).)))))))) ( -32.10, z-score = -1.42, R) >droEre2.scaffold_4784 9667872 108 - 25762168 AUGCAAGGCUCGAUAGUUUUCCAAGUGGCGGCAGCUCCACCUCAUUAUCCUUGUGGGCAACCACCAAGGAUGUGGUGAUCCCAGUGGUCGCAUCUCCACAUAUCAGCU ......((((.((((.........(((((.((.....((((....(((((((((((....))).)))))))).))))......)).))))).........)))))))) ( -37.07, z-score = -1.76, R) >droYak2.chr3L 7905932 108 - 24197627 AUUCAUGGCUCGAUAGUUUUCCAAGUGGCGGCAGCUCCACCUCAUUAUCCUUGUGGGCAACCACCAAGGAUGUGGUGAUCCCAGCGGUCGCAUCUCUACAUAUCAGCU ......((((.((((((......((..(((((.(((.((((....(((((((((((....))).)))))))).)))).....))).)))))..))..)).)))))))) ( -38.30, z-score = -2.66, R) >droSec1.super_0 9813010 108 + 21120651 AUACAUGGCUCGAUAGUUUUGCAAGUGGCGUCAGCUCCACCUCAUUAUCCUUGUGGGCAACCGCCAAGGAUGUGGCGAUCCCAGUGGUCGCAUCUCCACAUAUCAGCU ......((((.(((((..((((..((((.(....).))))..(((.((((((((((....))).))))))))))))))..)..((((........)))).)))))))) ( -38.10, z-score = -2.28, R) >droSim1.chr3L 17103569 108 + 22553184 AUACAUGGCUCGAUAGUUUUGCAAGUGGCGUCAGCUCCACCUCAUUAUCCUUGUGGGCAACCGCCAAGGAUGUGGUGAUCCCAGUGGUCGCAUCUCCACAUAUCAGCU ......((((.((((..........(((.((((...((((......((((((((((....))).))))))))))))))).)))((((........)))).)))))))) ( -36.10, z-score = -1.67, R) >consensus AUACAUGGCUCGAUAGUUUUCCAAGUGGCGUCAGCUCCACCUCAUUAUCCUUGUGGGCAACCGCCAAGGAUGUGGUGAUCCCAGUGGUCGCAUCUCCACAUAUCAGCU ......((((.((((.........((((.(....).))))..(((.((((((((((....))).))))))))))((((((.....)))))).........)))))))) (-33.12 = -32.76 + -0.36)

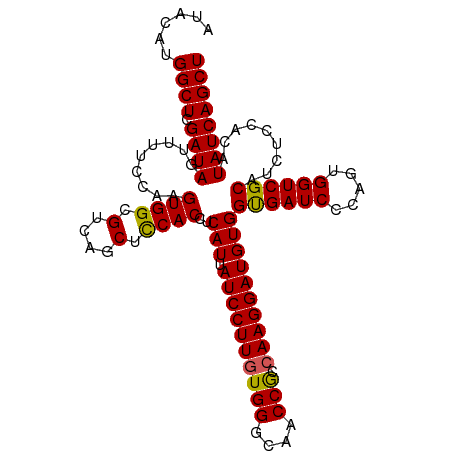
| Location | 17,765,954 – 17,766,064 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.12 |
| Shannon entropy | 0.32377 |
| G+C content | 0.52109 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -22.48 |
| Energy contribution | -23.73 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.558656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
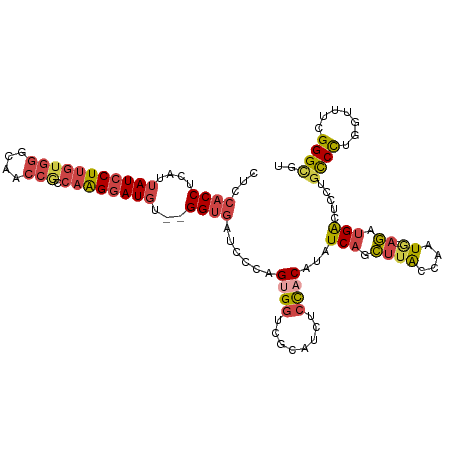
>dm3.chr3L 17765954 110 + 24543557 CUUCACCUCAUUAUCCUUUUGGGCAACCGCCAAGGAUGU--GGUGAUCCCAGUGGUCGCACCUCCACAUAUCAGCUUACCAAUA-AGAUGACUCCUGUCCUGGUUUCGGGCGU ..(((((....(((((((.(((....)))..))))))).--))))).(((.((((........))))..(((((((((....))-))..(((....))))))))...)))... ( -33.60, z-score = -1.40, R) >droAna3.scaffold_13337 14463836 108 + 23293914 ----ACCCUAUUAUCCUUGUGGGUAAAGUGCUGGAAUUUUUGGUCAUUCCAGUGGUCACAUCGGCCCAUAUCGUGUUGCAAAUU-ACAAGGUUUCGGUCUUCAUCUCGGGCGU ----.(((..((((((....))))))...((((((((........)))))))).........((((...(((.(((........-))).)))...))))........)))... ( -26.80, z-score = -0.42, R) >droEre2.scaffold_4784 9667906 109 - 25762168 CUCCACCUCAUUAUCCUUGUGGGCAACCACCAAGGAUGU--GGUGAUCCCAGUGGUCGCAUCUCCACAUAUCAGCUUACCAAUG-AGAUGACUCCUGCCCU-GUUUCGGGCGU ...((.((((((((((((((((....))).))))))))(--(((((.....((((........))))........)))))))))-)).))......((((.-.....)))).. ( -38.32, z-score = -2.43, R) >droYak2.chr3L 7905966 111 - 24197627 CUCCACCUCAUUAUCCUUGUGGGCAACCACCAAGGAUGU--GGUGAUCCCAGCGGUCGCAUCUCUACAUAUCAGCUUACCAAUGGAGCUGACUUCUGCCCUGGUUUCGGGCGU .((((((....(((((((((((....))).)))))))).--)))((..((((.(((.(........)...(((((((.......))))))).....)))))))..)))))... ( -39.90, z-score = -2.28, R) >droSec1.super_0 9813044 110 + 21120651 CUCCACCUCAUUAUCCUUGUGGGCAACCGCCAAGGAUGU--GGCGAUCCCAGUGGUCGCAUCUCCACAUAUCAGCUUACCAAUG-AGAUGACUCCUCCCCUUGUUUCGGGUGU ...((((((((.((((((((((....))).)))))))))--))(((..(.((.((...((((((...................)-)))))......)).)).)..))))))). ( -33.91, z-score = -1.34, R) >droSim1.chr3L 17103603 110 + 22553184 CUCCACCUCAUUAUCCUUGUGGGCAACCGCCAAGGAUGU--GGUGAUCCCAGUGGUCGCAUCUCCACAUAUCAGCUUACCAAUG-CGAUGACUCCUGCCCUUGUUUCGGGUGU ...((((....(((((((((((....))).)))))))).--)))).....(((.(((((((....................)))-)))).)))...((((.......)))).. ( -37.35, z-score = -2.13, R) >consensus CUCCACCUCAUUAUCCUUGUGGGCAACCGCCAAGGAUGU__GGUGAUCCCAGUGGUCGCAUCUCCACAUAUCAGCUUACCAAUG_AGAUGACUCCUGCCCUGGUUUCGGGCGU ...((((....(((((((((((....))).))))))))...))))......((((........)))).............................((((.......)))).. (-22.48 = -23.73 + 1.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:34 2011