| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,756,015 – 17,756,111 |
| Length | 96 |
| Max. P | 0.757436 |

| Location | 17,756,015 – 17,756,111 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 58.34 |
| Shannon entropy | 0.83323 |
| G+C content | 0.35602 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -0.65 |
| Energy contribution | -0.72 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.92 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.03 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757436 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
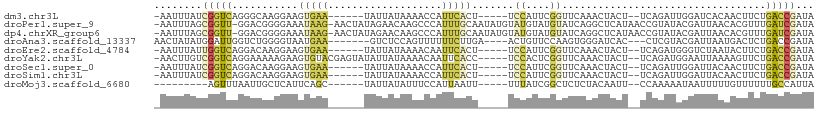
>dm3.chr3L 17756015 96 + 24543557 -AAUUUAUCGGUCAGGGCAAGGAAGUGAA------UAUUAUAAAACCAUUCACU-----UCCAUUCGGUUCAAACUACU--UCAGAUUGGAUCACAACUUCUGACCGAUA -....((((((((((((...(((((((((------(...........)))))))-----)))....(((((((.((...--..)).))))))).....)))))))))))) ( -35.90, z-score = -7.04, R) >droPer1.super_9 1576028 107 - 3637205 -AAUUUAGCGGUU-GGACGGGGAAAUAAG-AACUAUAGAACAAGCCCAUUUGCAAUAUGUAUGUAUGUAUCAGGCUCAUAACCGUAUACGAUUAACACGUUUGAUCGAUA -.......((((.-.((((.(..(((...-............((((....((((.(((....)))))))...))))......((....)))))..).))))..))))... ( -19.70, z-score = 0.48, R) >dp4.chrXR_group6 9546596 107 - 13314419 -AAUUUAGCGGUU-GGACGGGGAAAUAAG-AACUAUAGAACAAGCCCAUUUGCAAUAUGUAUGUAUGUAUCAGGCUCAUAACCGUAUACGAUUAACACGUUUGAUCGAUA -.......((((.-.((((.(..(((...-............((((....((((.(((....)))))))...))))......((....)))))..).))))..))))... ( -19.70, z-score = 0.48, R) >droAna3.scaffold_13337 14452974 96 + 23293914 AACUAUAUGGAUUGGUCUGGGGUAAUGAA-------GUCUCCAGUUUUUUCUUGA----ACUGUUCCAAGUGGGAUCAC---CUCGUACGAUUAAUGACUCUGACCGAUA ..........(((((((.(((((.((..(-------(((..((((((......))----))))......(((.((....---.)).))))))).)).)))))))))))). ( -24.10, z-score = -0.40, R) >droEre2.scaffold_4784 9657595 96 - 25762168 -AAUUUAUUGGUCAGGACAAGGAAGUGAA------UAUUAUAAAACAAUUCACU-----UCCAUUCGGUUCAAACUACU--UCAGAUGGGUCUAAUACUUCUGACCGAUA -....((((((((((((...(((((((((------(...........)))))))-----)))....((..((.......--.....))..))......)))))))))))) ( -30.20, z-score = -4.90, R) >droYak2.chr3L 7895382 102 - 24197627 -AACUUGUCGGUCAGGAAAAAGAAGUGUACGAGUAUAUUAUAAAACAAUUCACC-----UCCACUCGGUUCAAACUACU--UCAGAUGGAAUUAAAAGUUCUGACCGAUA -....((((((((((((......((((((....))))))...............-----((((((.(((.......)))--..)).))))........)))))))))))) ( -24.30, z-score = -2.00, R) >droSec1.super_0 9803135 96 + 21120651 -AAUUUAUCGGUCAGGACAAGGAAGUGAA------UAUUAUAAAACCAUUCACU-----UCCAUUCGGUUCAAACUACU--UCAGAUUGGAUUACAACUUCUGACCGAUA -....((((((((((((...(((((((((------(...........)))))))-----)))....(((((((.((...--..)).))))))).....)))))))))))) ( -34.20, z-score = -6.61, R) >droSim1.chr3L 17093058 96 + 22553184 -AAUUUAUCGGUCAGGACAAGGAAGUGAA------UAUUAUAAAACCAUUCACU-----UCCAUUCGGUUCAAACUACU--UCAGAUUGGAUUACAACUUCUGACCGAUA -....((((((((((((...(((((((((------(...........)))))))-----)))....(((((((.((...--..)).))))))).....)))))))))))) ( -34.20, z-score = -6.61, R) >droMoj3.scaffold_6680 19752009 88 - 24764193 ---------AGUUUAAUUGCUCAUUCAGC------UAUUAUAUUUCCAUUAAUU-----UUUAUCGGCUCUCUACAAUU--CCAAAAAUAAUUUUUGUUUUUUGCCAUUA ---------....((((.(((.....)))------.))))..............-----......(((.....((((..--.............)))).....))).... ( -8.76, z-score = -1.34, R) >consensus _AAUUUAUCGGUCAGGACAAGGAAGUGAA______UAUUAUAAAACCAUUCACU_____UCCAUUCGGUUCAAACUACU__UCAGAUAGGAUUAAAACUUCUGACCGAUA ........(((((...........(((((...................))))).......((....))..................................)))))... ( -0.65 = -0.72 + 0.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:31 2011