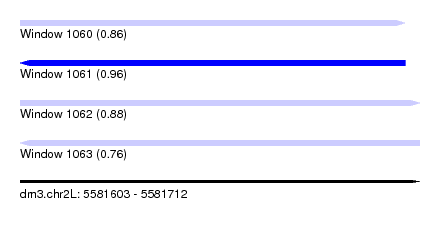
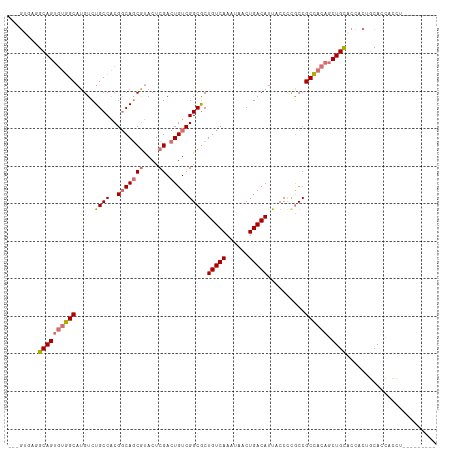
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,581,603 – 5,581,712 |
| Length | 109 |
| Max. P | 0.958021 |

| Location | 5,581,603 – 5,581,708 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.66 |
| Shannon entropy | 0.31983 |
| G+C content | 0.60217 |
| Mean single sequence MFE | -46.82 |
| Consensus MFE | -32.78 |
| Energy contribution | -34.40 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
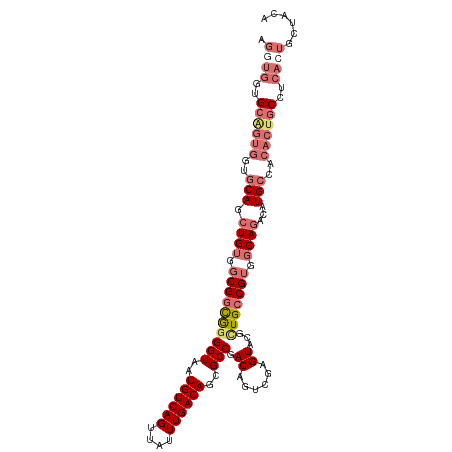
>dm3.chr2L 5581603 105 + 23011544 ---------AGGUGGUGCGGUGGUGCAGCUGUGGCGGCAAGGUAAUGUCAGUUAUUUGACAGCGCCGACAGUCGAGUACGCUGCCGUGGCAGACAUGCCACACUGCCUCACUGC ---------..(..(((.((..(((((((.(((.((((..(((..((((((....))))))..)))....))))..)))))))).((((((....))))))))..)).)))..) ( -50.80, z-score = -2.63, R) >droAna3.scaffold_12916 11029340 110 + 16180835 UAUCCAGCCAGCCCGUGGUGGGGCACAGGUGGUGCGGUGGGGUAAUGUCAGUUAUUUGACAGUGCCGACAGUCGAGUGUGUUGGCGGGGCAGACAUGCAA-GCUGCAAUGC--- (((((.(((..(((.....)))((((.....))))))).))))).((((((....)))))).((((((((........))))))))..((((.(......-))))).....--- ( -39.40, z-score = -0.07, R) >droSec1.super_5 3659459 105 + 5866729 ---------AGGUGGUGCAGUGGUGCAGCUGUGGCGGCGGGGUAAUGUCAGUUAUUUGACAGCGCCGACAGUCGAGUACGCUGCCGUGGCAGACAUGCCACACUGCCUCACUGC ---------.(((((.((((((..(((.((((.(((((((.(((.((((((....))))))....((.....))..))).))))))).))))...)))..)))))).))))).. ( -52.20, z-score = -2.76, R) >droSim1.chr2L 5376040 97 + 22036055 ---------AGGUGGUGCAGUGGUGCAGCUGUGGCGGCGGGGUAAUGUCAGUUAUUUGACAGCGCCGACAGUCGAGUACGCUGCCGUGGCAGACAUGCCACACUGC-------- ---------.......((((((..(((.((((.(((((((.(((.((((((....))))))....((.....))..))).))))))).))))...)))..))))))-------- ( -44.90, z-score = -2.11, R) >consensus _________AGGUGGUGCAGUGGUGCAGCUGUGGCGGCGGGGUAAUGUCAGUUAUUUGACAGCGCCGACAGUCGAGUACGCUGCCGUGGCAGACAUGCCACACUGCCUCAC___ ..............(((.(((((((((((.....((((..(((..((((((....))))))..)))....)))).....))))).((((((....)))))))))))).)))... (-32.78 = -34.40 + 1.62)

| Location | 5,581,603 – 5,581,708 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.66 |
| Shannon entropy | 0.31983 |
| G+C content | 0.60217 |
| Mean single sequence MFE | -39.85 |
| Consensus MFE | -26.88 |
| Energy contribution | -27.62 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5581603 105 - 23011544 GCAGUGAGGCAGUGUGGCAUGUCUGCCACGGCAGCGUACUCGACUGUCGGCGCUGUCAAAUAACUGACAUUACCUUGCCGCCACAGCUGCACCACCGCACCACCU--------- ((.(((..((((((((((......((..((((((((....)).))))))..))(((((......)))))..........)))))).))))..))).)).......--------- ( -43.00, z-score = -2.37, R) >droAna3.scaffold_12916 11029340 110 - 16180835 ---GCAUUGCAGC-UUGCAUGUCUGCCCCGCCAACACACUCGACUGUCGGCACUGUCAAAUAACUGACAUUACCCCACCGCACCACCUGUGCCCCACCACGGGCUGGCUGGAUA ---((...(((((-......).))))...(((.(((........))).)))..(((((......)))))..........)).(((((...((((......)))).)).)))... ( -30.20, z-score = -0.53, R) >droSec1.super_5 3659459 105 - 5866729 GCAGUGAGGCAGUGUGGCAUGUCUGCCACGGCAGCGUACUCGACUGUCGGCGCUGUCAAAUAACUGACAUUACCCCGCCGCCACAGCUGCACCACUGCACCACCU--------- ((((((..((((((((((...........(((((((....)).)))))((((.(((((......)))))......)))))))))).))))..)))))).......--------- ( -48.80, z-score = -4.07, R) >droSim1.chr2L 5376040 97 - 22036055 --------GCAGUGUGGCAUGUCUGCCACGGCAGCGUACUCGACUGUCGGCGCUGUCAAAUAACUGACAUUACCCCGCCGCCACAGCUGCACCACUGCACCACCU--------- --------(((((((((((....)))))).(((((((.....))((.(((((.(((((......)))))......))))).))..)))))...))))).......--------- ( -37.40, z-score = -2.58, R) >consensus ___GUGAGGCAGUGUGGCAUGUCUGCCACGGCAGCGUACUCGACUGUCGGCGCUGUCAAAUAACUGACAUUACCCCGCCGCCACAGCUGCACCACUGCACCACCU_________ ........((((((((((.....((((..(((((((....)).))))))))).(((((......)))))..........)))))).))))........................ (-26.88 = -27.62 + 0.75)

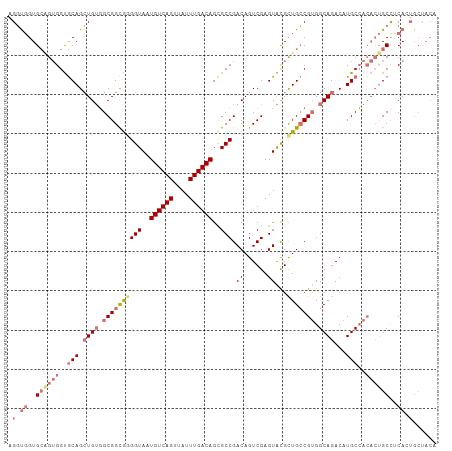
| Location | 5,581,603 – 5,581,712 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.57 |
| Shannon entropy | 0.28751 |
| G+C content | 0.59201 |
| Mean single sequence MFE | -48.77 |
| Consensus MFE | -33.25 |
| Energy contribution | -36.62 |
| Covariance contribution | 3.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5581603 109 + 23011544 AGGUGGUGCGGUGGUGCAGCUGUGGCGGCAAGGUAAUGUCAGUUAUUUGACAGCGCCGACAGUCGAGUACGCUGCCGUGGCAGACAUGCCACACUGCCUCACUGCUACA .((..(((.((..(((((((.(((.((((..(((..((((((....))))))..)))....))))..)))))))).((((((....))))))))..)).)))..))... ( -53.40, z-score = -2.90, R) >droAna3.scaffold_12916 11029349 105 + 16180835 AGCCCGUGGUGGGGCACAGGUGGUGCGGUGGGGUAAUGUCAGUUAUUUGACAGUGCCGACAGUCGAGUGUGUUGGCGGGGCAGACAUGCAA----GCUGCAAUGCCACA .((((......))))....((((((((((..((((.((((((....)))))).)))).........(((((((.((...)).)))))))..----)))))...))))). ( -40.90, z-score = -0.85, R) >droSec1.super_5 3659459 109 + 5866729 AGGUGGUGCAGUGGUGCAGCUGUGGCGGCGGGGUAAUGUCAGUUAUUUGACAGCGCCGACAGUCGAGUACGCUGCCGUGGCAGACAUGCCACACUGCCUCACUGCUACA .....(((((((((.((((....(((((((.(((..((((((....))))))..))).((......)).)))))))((((((....)))))).)))).))))))).)). ( -53.70, z-score = -2.70, R) >droSim1.chr2L 5376040 101 + 22036055 AGGUGGUGCAGUGGUGCAGCUGUGGCGGCGGGGUAAUGUCAGUUAUUUGACAGCGCCGACAGUCGAGUACGCUGCCGUGGCAGACAUGCCACACUGCUACA-------- .....((((((((..(((.((((.(((((((.(((.((((((....))))))....((.....))..))).))))))).))))...)))..)))))).)).-------- ( -47.10, z-score = -2.20, R) >consensus AGGUGGUGCAGUGGUGCAGCUGUGGCGGCGGGGUAAUGUCAGUUAUUUGACAGCGCCGACAGUCGAGUACGCUGCCGUGGCAGACAUGCCACACUGCCUCACUGCUACA .((((..((((((..(((.((((.((((((((((..((((((....))))))..))).((......))...))))))).))))...)))..))))))..))))...... (-33.25 = -36.62 + 3.38)

| Location | 5,581,603 – 5,581,712 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.57 |
| Shannon entropy | 0.28751 |
| G+C content | 0.59201 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -26.90 |
| Energy contribution | -29.65 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
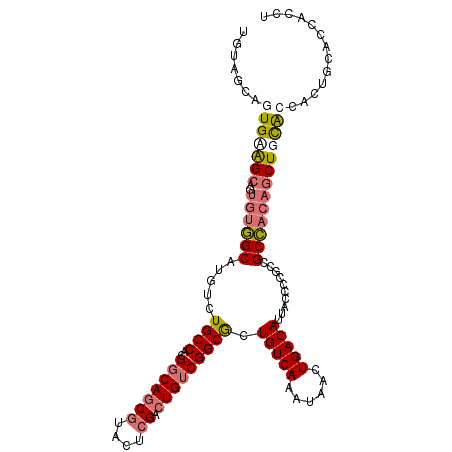
>dm3.chr2L 5581603 109 - 23011544 UGUAGCAGUGAGGCAGUGUGGCAUGUCUGCCACGGCAGCGUACUCGACUGUCGGCGCUGUCAAAUAACUGACAUUACCUUGCCGCCACAGCUGCACCACCGCACCACCU ....((.(((..((((((((((......((..((((((((....)).))))))..))(((((......)))))..........)))))).))))..))).))....... ( -43.20, z-score = -1.82, R) >droAna3.scaffold_12916 11029349 105 - 16180835 UGUGGCAUUGCAGC----UUGCAUGUCUGCCCCGCCAACACACUCGACUGUCGGCACUGUCAAAUAACUGACAUUACCCCACCGCACCACCUGUGCCCCACCACGGGCU ...(((((.((...----..))))))).((((.(((.(((........))).)))..(((((......)))))..........((((.....))))........)))). ( -28.20, z-score = -0.55, R) >droSec1.super_5 3659459 109 - 5866729 UGUAGCAGUGAGGCAGUGUGGCAUGUCUGCCACGGCAGCGUACUCGACUGUCGGCGCUGUCAAAUAACUGACAUUACCCCGCCGCCACAGCUGCACCACUGCACCACCU ....((((((..((((((((((...........(((((((....)).)))))((((.(((((......)))))......)))))))))).))))..))))))....... ( -49.00, z-score = -3.47, R) >droSim1.chr2L 5376040 101 - 22036055 --------UGUAGCAGUGUGGCAUGUCUGCCACGGCAGCGUACUCGACUGUCGGCGCUGUCAAAUAACUGACAUUACCCCGCCGCCACAGCUGCACCACUGCACCACCU --------....(((((((((((....)))))).(((((((.....))((.(((((.(((((......)))))......))))).))..)))))...)))))....... ( -37.60, z-score = -1.89, R) >consensus UGUAGCAGUGAAGCAGUGUGGCAUGUCUGCCACGGCAGCGUACUCGACUGUCGGCGCUGUCAAAUAACUGACAUUACCCCGCCGCCACAGCUGCACCACUGCACCACCU ....((.(((..((((((((((.....((((..(((((((....)).))))))))).(((((......)))))..........)))))).))))..))).))....... (-26.90 = -29.65 + 2.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:28 2011