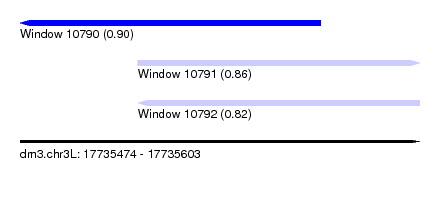
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,735,474 – 17,735,603 |
| Length | 129 |
| Max. P | 0.900048 |

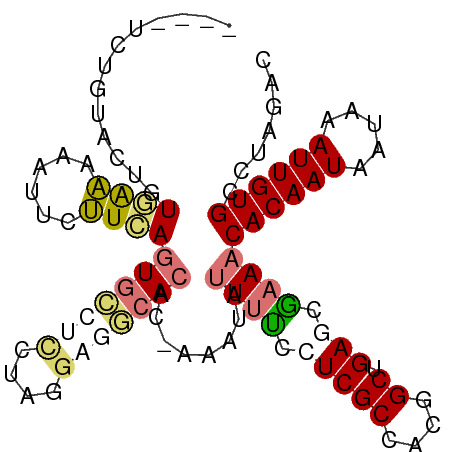
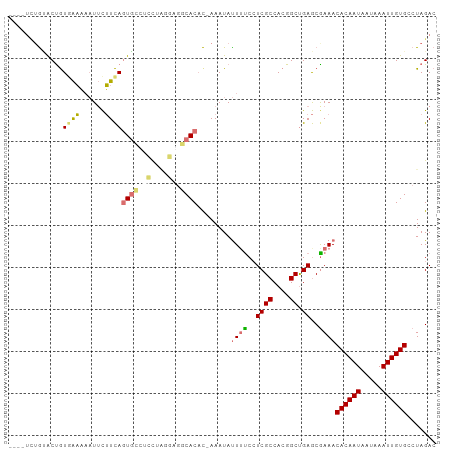
| Location | 17,735,474 – 17,735,571 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 73.53 |
| Shannon entropy | 0.42969 |
| G+C content | 0.41354 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -12.96 |
| Energy contribution | -14.65 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
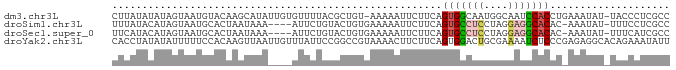
>dm3.chr3L 17735474 97 - 24543557 ----UUUACGCUGU-AAAAAUUCUUCAGUGGCAAUGGCAAUCCACCUGAAAUAUUACCCUCGCCACGGCUGAGCGAAACACAAUAAUAAAUUGUGCCUAGAC ----....((((..-...(((..((((((((..........))).)))))..)))......((....))..))))...((((((.....))))))....... ( -17.90, z-score = -0.50, R) >droSim1.chr3L 17069665 97 - 22553184 ----UCUGUACUGUGAAAAAUUCUUCAGUGCCUCCUAGGAGGCACAC-AAAUAUUUUCCUCGCCACGGCUGAGCGAAACACAAUAAUAAAUUGUGCCUAGAC ----..(((....((((......))))(((((((....)))))))))-).....(((((((((....)).))).))))((((((.....))))))....... ( -28.20, z-score = -3.54, R) >droSec1.super_0 9779937 97 - 21120651 ----UCUGUACUGUGAAAAAUUCUUCAGUGCCUCCUAGGAGGCACAC-AAAUAUUUUCAUCGCCACGGCUGAGCAAAACACAAUAAUAAAUUGUGCCUAGAC ----..(((...(((((((........(((((((....)))))))..-.....))))))).((....))...)))...((((((.....))))))....... ( -24.36, z-score = -2.43, R) >droEre2.scaffold_4784 9637135 101 + 25762168 CCAAAGAGAACCAUAGCCUUUAUGUUAUUUAGUUUAUUGUGGCACAC-AAAUACUUUCCUCGCCACGGCCGAACGAAACACAAUAAUAAAUUGUGACUAGAC ............(((((......)))))...((((.(((((((....-.............)))))))..))))....((((((.....))))))....... ( -16.13, z-score = -0.72, R) >consensus ____UCUGUACUGUGAAAAAUUCUUCAGUGCCUCCUAGGAGGCACAC_AAAUAUUUUCCUCGCCACGGCUGAGCGAAACACAAUAAUAAAUUGUGCCUAGAC .............((((......))))(((((((....)))))))........((((.(((((....)).))).))))((((((.....))))))....... (-12.96 = -14.65 + 1.69)

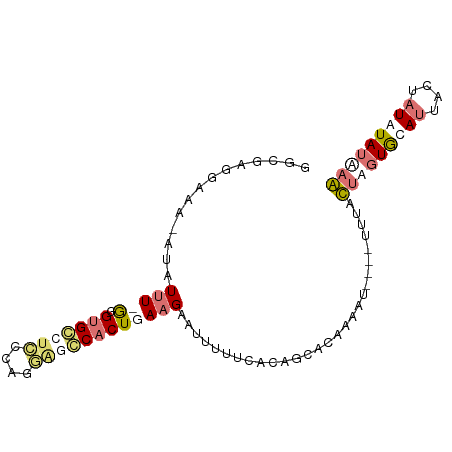
| Location | 17,735,512 – 17,735,603 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 59.27 |
| Shannon entropy | 0.65215 |
| G+C content | 0.36559 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -7.06 |
| Energy contribution | -9.50 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.863242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17735512 91 + 24543557 GGCGAGGGUA-AUAUUUCAGGUGGAUUGCCAUUGCCACUGAAGAAUUUUU-ACAGCGUAAAACACAAUAUGCUUGUACAUUACUAUAUAUAAG (((((.((((-((...........)))))).))))).............(-((((((((........))))).))))................ ( -19.10, z-score = -1.11, R) >droSim1.chr3L 17069703 87 + 22553184 GGCGAGGAAA-AUAUUU-GUGUGCCUCCUAGGAGGCACUGAAGAAUUUUUCACAGUACAGAAU----UUUAUUAGUGCAUUACUAUGUAUAAA .((...((((-(.((((-..(((((((....)))))))....)))))))))...)).......----.......((((((....))))))... ( -23.50, z-score = -2.69, R) >droSec1.super_0 9779975 87 + 21120651 GGCGAUGAAA-AUAUUU-GUGUGCCUCCUAGGAGGCACUGAAGAAUUUUUCACAGUACAGAAU----UUUAUUAGUGCAUUACUAUGUAUGAA .((..(((((-(.((((-..(((((((....)))))))....))))))))))..)).......----.......((((((....))))))... ( -24.60, z-score = -3.10, R) >droYak2.chr3L 7874516 93 - 24197627 AAUAUUUCUGUGCCUCUCGGGAGAUUUUCGCAGUCCACUGAAGAAGUUUUACGGCCGGAAUAAACAAUUAACUUGUGGAAAAAUAUAUAGGUG .....(((((.(((......((((((((..(((....)))..))))))))..))))))))..........(((((((........))))))). ( -21.10, z-score = -1.04, R) >consensus GGCGAGGAAA_AUAUUU_GGGUGCCUCCCAGGAGCCACUGAAGAAUUUUUCACAGCACAAAAU____UUUACUAGUGCAUUACUAUAUAUAAA ..............(((((.(((((((....))))))))))))............................(((((((((....))))))))) ( -7.06 = -9.50 + 2.44)

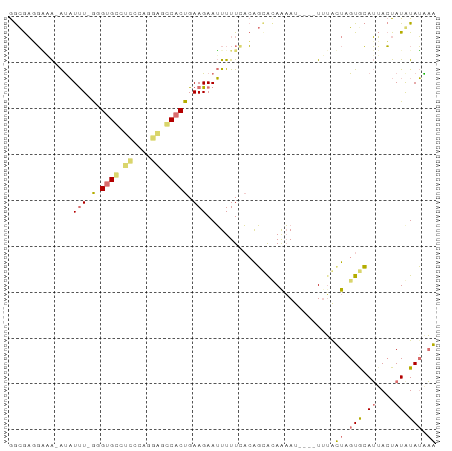
| Location | 17,735,512 – 17,735,603 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 59.27 |
| Shannon entropy | 0.65215 |
| G+C content | 0.36559 |
| Mean single sequence MFE | -15.98 |
| Consensus MFE | -6.99 |
| Energy contribution | -7.67 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.819708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
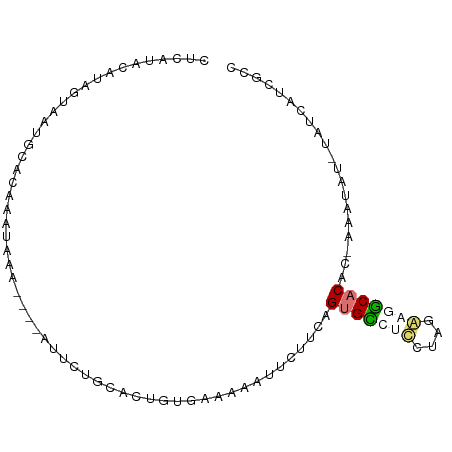
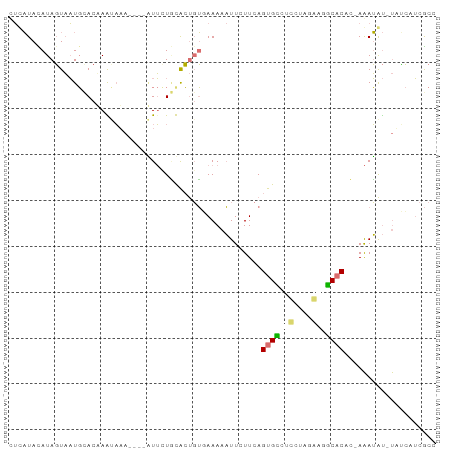
>dm3.chr3L 17735512 91 - 24543557 CUUAUAUAUAGUAAUGUACAAGCAUAUUGUGUUUUACGCUGU-AAAAAUUCUUCAGUGGCAAUGGCAAUCCACCUGAAAUAU-UACCCUCGCC ....((((..((((..(((((.....)))))..))))..)))-)..(((..((((((((..........))).)))))..))-)......... ( -13.60, z-score = 0.27, R) >droSim1.chr3L 17069703 87 - 22553184 UUUAUACAUAGUAAUGCACUAAUAAA----AUUCUGUACUGUGAAAAAUUCUUCAGUGCCUCCUAGGAGGCACAC-AAAUAU-UUUCCUCGCC ......(((((((.............----......)))))))............(((((((....)))))))..-......-.......... ( -19.51, z-score = -3.11, R) >droSec1.super_0 9779975 87 - 21120651 UUCAUACAUAGUAAUGCACUAAUAAA----AUUCUGUACUGUGAAAAAUUCUUCAGUGCCUCCUAGGAGGCACAC-AAAUAU-UUUCAUCGCC ......(((((((.............----......)))))))............(((((((....)))))))..-......-.......... ( -19.51, z-score = -3.16, R) >droYak2.chr3L 7874516 93 + 24197627 CACCUAUAUAUUUUUCCACAAGUUAAUUGUUUAUUCCGGCCGUAAAACUUCUUCAGUGGACUGCGAAAAUCUCCCGAGAGGCACAGAAAUAUU .......((((((((..((((.....))))........(((.......(((..(((....))).)))..(((....))))))..)))))))). ( -11.30, z-score = 1.13, R) >consensus CUCAUACAUAGUAAUGCACAAAUAAA____AUUCUGCACUGUGAAAAAUUCUUCAGUGCCUCCUAGAAGGCACAC_AAAUAU_UAUCAUCGCC .......................................................(((((((....))))))).................... ( -6.99 = -7.67 + 0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:29 2011