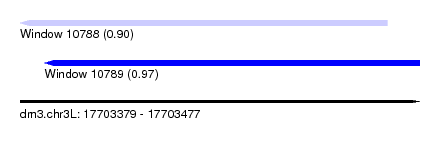
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,703,379 – 17,703,477 |
| Length | 98 |
| Max. P | 0.965895 |

| Location | 17,703,379 – 17,703,469 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.90 |
| Shannon entropy | 0.60350 |
| G+C content | 0.34727 |
| Mean single sequence MFE | -21.09 |
| Consensus MFE | -10.53 |
| Energy contribution | -10.25 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.895924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
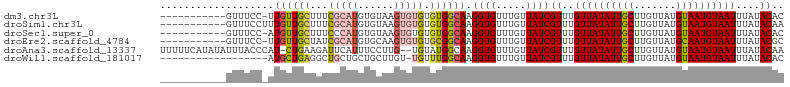
>dm3.chr3L 17703379 90 - 24543557 -GUUGCUUUCGCAUGUGUAAG--------------UG-UGUGUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGUAAUGUAAUUUAUACACACCCAC-- -.(((((..((((..(....)--------------..-)))).)))))(((((.(((........((((((((((.......))))))))))..))).)))))...-- ( -23.90, z-score = -1.76, R) >droSim1.chr3L 17042984 91 - 22553184 UGUUGCUUUCGCAUGUGUAAG--------------UG-UGUGUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGUAAUGUAAUUUAUACAAACCCAU-- (((..(...(((((......)--------------))-)).)..))).((.((((((....((..((((((((((.......))))))))))..))))))))))..-- ( -26.60, z-score = -3.60, R) >droSec1.super_0 9754961 90 - 21120651 -GUUGCUUUCCCAUGUGUAAG--------------UG-UGUGUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGUAAUGUAAUUUAUACACACCCAU-- -.((((..........))))(--------------((-(((((((...((((.....))))....((((((((((.......))))))))))))))))))))....-- ( -20.40, z-score = -1.21, R) >droYak2.chr3L 7849428 104 + 24197627 -GUUGCUUUCGCAUCUGGUAGUGUGUGUGCCUGUAUG-UGUGUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGUAAUGUAAUUUAUACACACCCAU-- -.(((((..((((((.((((.......)))).).)))-))...)))))(((((.(((........((((((((((.......))))))))))..))).)))))...-- ( -27.40, z-score = -1.49, R) >droEre2.scaffold_4784 9612606 90 + 25762168 -GUUGCUAUCGCAUGUGCAAG--------------UG-UGUGCGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGCAAUGUAAUUUAUACGCACCCAU-- -(((((...(((((......)--------------))-)).)))))..(((((.(((........((((((((((.......))))))))))..))).)))))...-- ( -26.90, z-score = -1.34, R) >droAna3.scaffold_13337 14406622 99 - 23293914 --AUAUUUACCCAUCUGAAGAUUCAUUUCCU----UG-UGUAUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGUAAUGUAAUUUAUACAAGCCCAU-- --..........................(((----((-(.....)))))).((((((....((..((((((((((.......))))))))))..))))))))....-- ( -18.70, z-score = -1.17, R) >droWil1.scaffold_181017 608322 83 + 939937 -------------------GCUGCUGCUGCU----UGUUGUUUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGUAAUGUAAUUUAUACACACGCAU-- -------------------((....))..((----((((....))))))((((.(((....((..((((((((((.......))))))))))..))))).))))..-- ( -19.40, z-score = -0.45, R) >droVir3.scaffold_13049 18504124 82 + 25233164 --------------------------UAUUUUUAUAUUUUCUUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUGUGUAAUGUAAUUUAUACACACACACAC --------------------------.......................((((.(((....((..((((((((((.......))))))))))..))..))).)))).. ( -14.30, z-score = -0.61, R) >droMoj3.scaffold_6680 19706177 83 + 24764193 -------------------------UUUUUUUUGGCGAGUGCAGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUGUGUAACGUAAUUUAUAUAUAUACACGA -------------------------........(((((..((((((.....))))))..))))).(((((((........)))))))..................... ( -14.60, z-score = 0.23, R) >droGri2.scaffold_15110 6060269 87 - 24565398 ---------------------CUUCUUUUUUUGCACACUCCCUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUGUGUAAUGUAAUUUAUAUACACACACAC ---------------------........(((((.((.....)))))))((((.(((....((..((((((((((.......))))))))))..))..))).)))).. ( -18.70, z-score = -1.73, R) >consensus __UUGCUUUC_CAU_UG_AAG______U__U____UG_UGUGUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGUAAUGUAAUUUAUACACACCCAU__ .................................................(((..(((....((..((((((((((.......))))))))))..))..)))..))).. (-10.53 = -10.25 + -0.28)
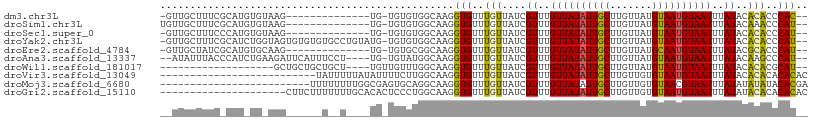
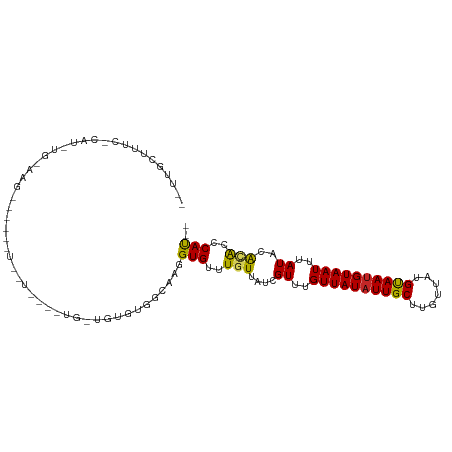
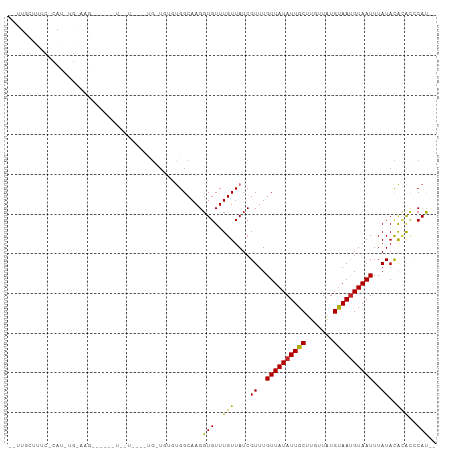
| Location | 17,703,385 – 17,703,477 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Shannon entropy | 0.33510 |
| G+C content | 0.34370 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -14.88 |
| Energy contribution | -15.43 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.965895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
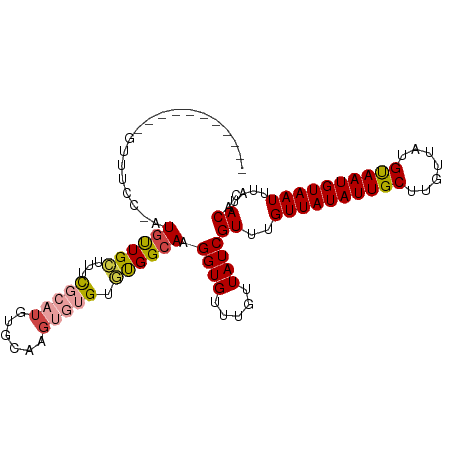
>dm3.chr3L 17703385 92 - 24543557 -----------GUUUCC-UUGUUGCUUUCGCAUGUGUAAGUGUGUGUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGUAAUGUAAUUUAUACAC -----------....((-((((..(...(((((......))))).)..))))))...........((..((((((((((.......))))))))))....)).. ( -25.80, z-score = -4.14, R) >droSim1.chr3L 17042990 93 - 22553184 -----------GUUUCCUUUGUUGCUUUCGCAUGUGUAAGUGUGUGUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGUAAUGUAAUUUAUACAA -----------.....((((((..(...(((((......))))).)..))))))...........((..((((((((((.......))))))))))....)).. ( -23.30, z-score = -3.54, R) >droSec1.super_0 9754967 92 - 21120651 -----------GUUUCC-AUGUUGCUUUCCCAUGUGUAAGUGUGUGUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGUAAUGUAAUUUAUACAC -----------.....(-((.(((((..(.((..(....)..)).).))))).))).........((..((((((((((.......))))))))))....)).. ( -19.00, z-score = -1.48, R) >droEre2.scaffold_4784 9612612 92 + 25762168 -----------GUUUCC-UUGUUGCUAUCGCAUGUGCAAGUGUGUGCGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGCAAUGUAAUUUAUACGC -----------....((-(((((((...(((((......))))).)))))))))..........(((..((((((((((.......))))))))))....))). ( -30.10, z-score = -3.56, R) >droAna3.scaffold_13337 14406628 101 - 23293914 UUUUUCAUAUAUUUACCCAU-CUGAAGAUUCAUUUCCUUG--UGUAUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGUAAUGUAAUUUAUACAA ....................-.....(((.((...(((((--(.....))))))....))..)))((..((((((((((.......))))))))))....)).. ( -17.90, z-score = -1.39, R) >droWil1.scaffold_181017 608328 85 + 939937 ------------------AUGCUGAGGCUGCUGCUGCUUGU-UGUUUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGUAAUGUAAUUUAUACAC ------------------.(((..((((.((....))..))-..))..))).((((.....))))((..((((((((((.......))))))))))....)).. ( -17.60, z-score = -0.15, R) >consensus ___________GUUUCC_AUGUUGCUUUCGCAUGUGCAAGUGUGUGUGGCAAGGUGUUUGUUAUCGUUUGUUAUAUUGCUUGUUAUGUAAUGUAAUUUAUACAC ...................((((((...(((((......))))).)))))).((((.....))))((..((((((((((.......))))))))))....)).. (-14.88 = -15.43 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:27 2011