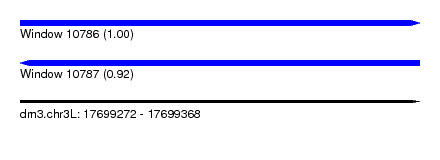
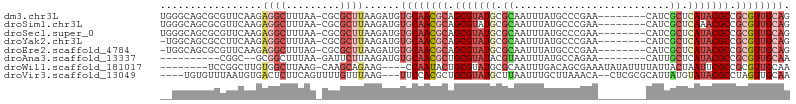
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,699,272 – 17,699,368 |
| Length | 96 |
| Max. P | 0.999180 |

| Location | 17,699,272 – 17,699,368 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 71.06 |
| Shannon entropy | 0.57317 |
| G+C content | 0.50724 |
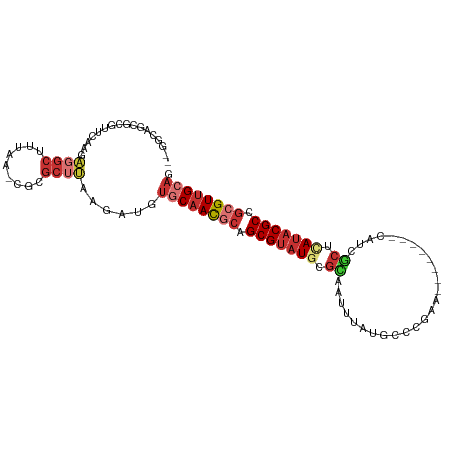
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -22.43 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17699272 96 + 24543557 CUGCAACGCGGCGUAUGAGCGAUG--------UUCGGGCAUAAAUUGCGCAUACGCUGCGUUGCACAUCUUAAGCGCG-UUAAAGCCUCUUGAACGCGCUGCCCA .((((((((((((((((.(((((.--------...........))))).)))))))))))))))).......((((((-((.(((...))).))))))))..... ( -47.50, z-score = -4.55, R) >droSim1.chr3L 17038759 96 + 22553184 CUGCAACGCGGCGUUUGAGCGAUG--------UUCGGGCAUAAAUUGCGCAUACGCUGCGUUGCACAUCUUAAGCGCG-UUAAAGCCUCUUGAACGCGCUGCCCA .(((((((((((((.((.(((((.--------...........))))).)).))))))))))))).......((((((-((.(((...))).))))))))..... ( -43.60, z-score = -3.27, R) >droSec1.super_0 9745228 96 + 21120651 CUGCAACGCGGCGUAUGAGCGAUG--------UUCGGGCAUAAAUUGCGCAUACGCUGCGUUGCACAUCUUAAGCGCG-UUAAAGCCUCUUGAACGCGCUGCCCA .((((((((((((((((.(((((.--------...........))))).)))))))))))))))).......((((((-((.(((...))).))))))))..... ( -47.50, z-score = -4.55, R) >droYak2.chr3L 7844978 95 - 24197627 CUGCAACGCGGCGUAUGAGCGAUG--------UUCGGGCAUAAAUUGCGCAUACGCUGCGUUGCACAUCUUAAGCGCG-UUAAAGCCUCUUGAAGGCGCUGCCA- .((((((((((((((((.(((((.--------...........))))).)))))))))))))))).......(((((.-((.(((...))).)).)))))....- ( -43.90, z-score = -3.18, R) >droEre2.scaffold_4784 9608280 95 - 25762168 CUGCAACGCGGCGUAUGAGCGAUG--------UUCGGGCAUAAAUUGCGCAUACGCUGCGUUGCACAUCUUAAGCGCG-CUAAAGCCUCUUGAACGCGCUGCCA- .((((((((((((((((.(((((.--------...........))))).)))))))))))))))).......((((((-...(((...)))...))))))....- ( -44.50, z-score = -3.41, R) >droAna3.scaffold_13337 14402587 84 + 23293914 UUGCAACGCGGCGUAUGAGCAAUG--------UUCUGGCAUAAAUUACGUAUACGCAGCGUUGCACAUCUUAAGAAUC-UUAAAGCCGC--GCCG---------- .((((((((.(((((((....(((--------(....))))........))))))).)))))))).............-..........--....---------- ( -23.40, z-score = -0.42, R) >droWil1.scaffold_181017 600238 92 - 939937 UUGCAACGCGGCGAAUUAGUAAUAAAAUAUAUUUCGCUGUCAAAUUGCGCAUACGCAGUAUUGG----CUUCUGCUUG-CUUAAGCCACAAGCCGGA-------- .......((((((((.((...........)).))))))))...((((((....)))))).((((----(((..((((.-...))))...))))))).-------- ( -25.70, z-score = -0.75, R) >droVir3.scaffold_13049 18499538 96 - 25233164 UUGCAACUAGGCGUAUACAUAAUGCGCGAG--UGUUUAAGCAAAUUAAGCAUACGCAGCGUGGAA---CUUAAACAAAACUGAAGAGUCACAUUAAACACA---- ..((......((((.......))))(((.(--(((((((.....)))))))).))).))((((..---(((...........)))..))))..........---- ( -20.70, z-score = -0.86, R) >consensus CUGCAACGCGGCGUAUGAGCGAUG________UUCGGGCAUAAAUUGCGCAUACGCUGCGUUGCACAUCUUAAGCGCG_UUAAAGCCUCUUGAACGCGCUGCC__ .((((((((.(((((((.(((((....................))))).))))))).))))))))........................................ (-22.43 = -23.40 + 0.97)
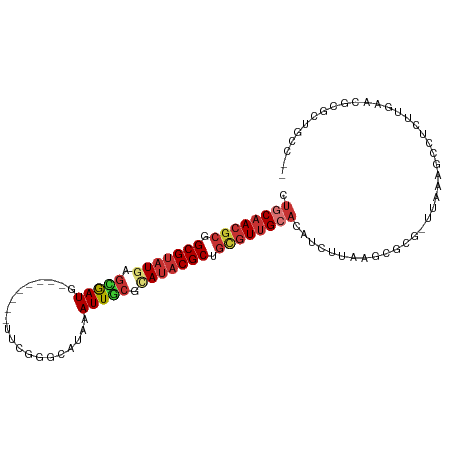
| Location | 17,699,272 – 17,699,368 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 71.06 |
| Shannon entropy | 0.57317 |
| G+C content | 0.50724 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -18.46 |
| Energy contribution | -19.65 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.915331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17699272 96 - 24543557 UGGGCAGCGCGUUCAAGAGGCUUUAA-CGCGCUUAAGAUGUGCAACGCAGCGUAUGCGCAAUUUAUGCCCGAA--------CAUCGCUCAUACGCCGCGUUGCAG .....((((((((.(((...))).))-)))))).......((((((((.(((((((.((..............--------....)).))))))).)))))))). ( -39.17, z-score = -2.02, R) >droSim1.chr3L 17038759 96 - 22553184 UGGGCAGCGCGUUCAAGAGGCUUUAA-CGCGCUUAAGAUGUGCAACGCAGCGUAUGCGCAAUUUAUGCCCGAA--------CAUCGCUCAAACGCCGCGUUGCAG .....((((((((.(((...))).))-)))))).......((((((((.((((.((.((..............--------....)).)).)))).)))))))). ( -35.07, z-score = -0.68, R) >droSec1.super_0 9745228 96 - 21120651 UGGGCAGCGCGUUCAAGAGGCUUUAA-CGCGCUUAAGAUGUGCAACGCAGCGUAUGCGCAAUUUAUGCCCGAA--------CAUCGCUCAUACGCCGCGUUGCAG .....((((((((.(((...))).))-)))))).......((((((((.(((((((.((..............--------....)).))))))).)))))))). ( -39.17, z-score = -2.02, R) >droYak2.chr3L 7844978 95 + 24197627 -UGGCAGCGCCUUCAAGAGGCUUUAA-CGCGCUUAAGAUGUGCAACGCAGCGUAUGCGCAAUUUAUGCCCGAA--------CAUCGCUCAUACGCCGCGUUGCAG -.(((.(((((((...))))).....-.))))).......((((((((.(((((((.((..............--------....)).))))))).)))))))). ( -33.67, z-score = -0.96, R) >droEre2.scaffold_4784 9608280 95 + 25762168 -UGGCAGCGCGUUCAAGAGGCUUUAG-CGCGCUUAAGAUGUGCAACGCAGCGUAUGCGCAAUUUAUGCCCGAA--------CAUCGCUCAUACGCCGCGUUGCAG -....((((((((.(((...))).))-)))))).......((((((((.(((((((.((..............--------....)).))))))).)))))))). ( -39.17, z-score = -1.75, R) >droAna3.scaffold_13337 14402587 84 - 23293914 ----------CGGC--GCGGCUUUAA-GAUUCUUAAGAUGUGCAACGCUGCGUAUACGUAAUUUAUGCCAGAA--------CAUUGCUCAUACGCCGCGUUGCAA ----------((((--((((((((((-(...))))))(((.((((..(((.(((((.......))))))))..--------..)))).)))..)))))))))... ( -28.90, z-score = -1.98, R) >droWil1.scaffold_181017 600238 92 + 939937 --------UCCGGCUUGUGGCUUAAG-CAAGCAGAAG----CCAAUACUGCGUAUGCGCAAUUUGACAGCGAAAUAUAUUUUAUUACUAAUUCGCCGCGUUGCAA --------...(((((.((.(((...-.))))).)))----)).....((((.(((((..........(((((.((((......)).)).)))))))))))))). ( -23.00, z-score = 0.06, R) >droVir3.scaffold_13049 18499538 96 + 25233164 ----UGUGUUUAAUGUGACUCUUCAGUUUUGUUUAAG---UUCCACGCUGCGUAUGCUUAAUUUGCUUAAACA--CUCGCGCAUUAUGUAUACGCCUAGUUGCAA ----.........((..(((...............((---(.....)))((((((((.((((.(((.......--...))).)))).))))))))..)))..)). ( -20.60, z-score = -0.53, R) >consensus __GGCAGCGCGUUCAAGAGGCUUUAA_CGCGCUUAAGAUGUGCAACGCAGCGUAUGCGCAAUUUAUGCCCGAA________CAUCGCUCAUACGCCGCGUUGCAG ........................................((((((((.(((((((.((..........................)).))))))).)))))))). (-18.46 = -19.65 + 1.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:26 2011