| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,678,535 – 17,678,662 |
| Length | 127 |
| Max. P | 0.999307 |

| Location | 17,678,535 – 17,678,631 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 72.76 |
| Shannon entropy | 0.51457 |
| G+C content | 0.38062 |
| Mean single sequence MFE | -23.71 |
| Consensus MFE | -14.41 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17678535 96 - 24543557 GAAUGCCGGAAAAUCGCUGGCAUUUAUGCUCGCGAA--UAUUUUCGGUAGUGCCUGCC-UCCUCACUUAUUUAUAUGUAUUUCAUUUGAAUCGG----UAUGU- ..(((((((((((((((.((((....)))).)))).--..)))))(((((...)))))-.........(((((.(((.....))).))))))))----)))..- ( -25.60, z-score = -2.01, R) >droEre2.scaffold_4784 9588415 85 + 25762168 -AAUGCCAGAAAAUCGCUGGCAUUUAUGCUCGCGAAAUUGUUUUUGGUAG-----------CUCACUUAUUUACUCGU--UUCAUUUGAAUCGG----UAUGU- -..(((((((((.((((.((((....)))).)))).....))))))))).-----------..........(((.((.--(((....))).)))----))...- ( -22.50, z-score = -2.39, R) >droYak2.chr3L 7824188 100 + 24197627 GAAUGCCGGAAAAUCGCUGGCAUUUAUGCUCGCGAA--UAUUUUCGGUGGUGCUUGUC-UGCUCACUUAUUUAUAUGUGUUUCAUUUGAAUCGGUCGCUAUGU- ....((((((((.((((.((((....)))).)))).--..))))))))(((((.....-.))..(((.(((((.(((.....))).))))).))).)))....- ( -25.00, z-score = -0.92, R) >droSec1.super_0 9724869 96 - 21120651 GAAUGCCGGAAAAUUGCUGGCAUUUAUGCUCGCGAA--UAUUUUCGGUAUUGCCUGCC-UCCUCACUUAUUUAUAUGUGUUUCAUUUGAAUCGG----UAUGU- .(((((((((((.((((.((((....)))).)))).--..)))))))))))((.((((-....(((..........))).(((....)))..))----)).))- ( -24.70, z-score = -1.91, R) >droSim1.chr3L 17017568 96 - 22553184 GAAUGCCGGAAAAUCGCUGGCAUUUAUGCUCGCGAA--UAUUUUCGGUAGUGCCUGCC-UCCUCACGUAUUUAUAUGUGUUUCAUUUGAAUCGG----UAUGU- ...(((((((((.((((.((((....)))).)))).--..)))))))))(((((....-....((((((....)))))).(((....)))..))----)))..- ( -28.90, z-score = -2.56, R) >droGri2.scaffold_15110 6037489 93 - 24565398 -------AAAAAACCAUUUUCAUAUAUGAUGACUGUUUAUUUACUUACAAUUCAUGCCAUAUAUAUAUAUAUAAAUGGCGUUCAUUUAUUUCAG----UCGGUC -------.....(((....(((....))).(((((..................(((((((.((((....)))).)))))))..........)))----))))). ( -15.59, z-score = -1.72, R) >consensus GAAUGCCGGAAAAUCGCUGGCAUUUAUGCUCGCGAA__UAUUUUCGGUAGUGCCUGCC_UCCUCACUUAUUUAUAUGUGUUUCAUUUGAAUCGG____UAUGU_ ...(((((((((.((((.((((....)))).)))).....)))))))))................((.(((((.(((.....))).))))).)).......... (-14.41 = -14.70 + 0.29)


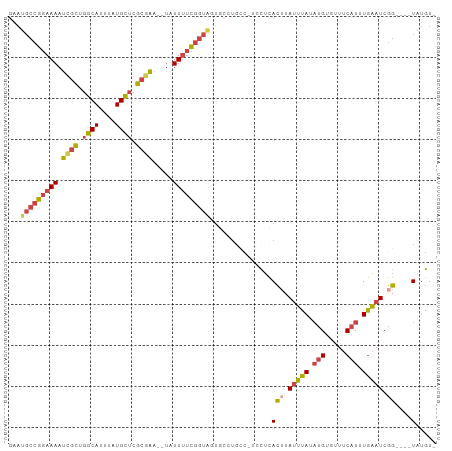
| Location | 17,678,558 – 17,678,662 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.06 |
| Shannon entropy | 0.32993 |
| G+C content | 0.50816 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -18.29 |
| Energy contribution | -20.73 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17678558 104 + 24543557 AUAUAAAUAAGUGAGGAGGCAGGCACUACCGAAAAUAUUCGCGAGCAUAAAUGCCAGCGAUUUUCCGGCAUUCAUUCGCAGCCUCCCCGAGUUGCCGCCUCCCC--------- ..............((((((.((((((...(((((...((((..(((....)))..))))))))).(((...........)))......)).))))))))))..--------- ( -32.70, z-score = -2.69, R) >droYak2.chr3L 7824215 95 - 24197627 AUAUAAAUAAGUGAGCAGACAAGCACCACCGAAAAUAUUCGCGAGCAUAAAUGCCAGCGAUUUUCCGGCAUUCACUAGGCGCCUCCCCUUUCCCU------------------ .........(((((((......))....(((.(((...((((..(((....)))..)))).))).)))...)))))...................------------------ ( -19.40, z-score = -1.26, R) >droSec1.super_0 9724892 112 + 21120651 AUAUAAAUAAGUGAGGAGGCAGGCAAUACCGAAAAUAUUCGCGAGCAUAAAUGCCAGCAAUUUUCCGGCAUUCAUUCGCAGCAUCCCCGUGUUGCAACCCGCCCCGCAACAG- ..........(((.((.((...(((((((.(((....)))(((((....((((((...........))))))..))))).........)))))))...)).)).))).....- ( -30.90, z-score = -2.18, R) >droSim1.chr3L 17017591 113 + 22553184 AUAUAAAUACGUGAGGAGGCAGGCACUACCGAAAAUAUUCGCGAGCAUAAAUGCCAGCGAUUUUCCGGCAUUCAUUCGCAGCCUCCCCGUGUUGCAACCCGCCCCCCCCACCG .....((((((.(.((((((.((.....))..........(((((....((((((...........))))))..))))).)))))))))))))((.....))........... ( -32.10, z-score = -2.46, R) >consensus AUAUAAAUAAGUGAGGAGGCAGGCACUACCGAAAAUAUUCGCGAGCAUAAAUGCCAGCGAUUUUCCGGCAUUCAUUCGCAGCCUCCCCGUGUUGCAACCCGCCC_________ .....((((((.(.((((((..........(((....)))(((((....((((((...........))))))..))))).))))))))))))).................... (-18.29 = -20.73 + 2.44)

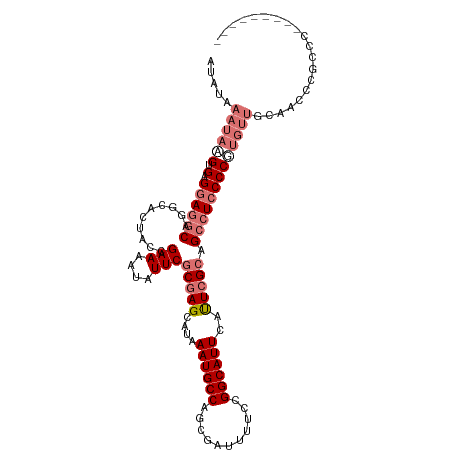
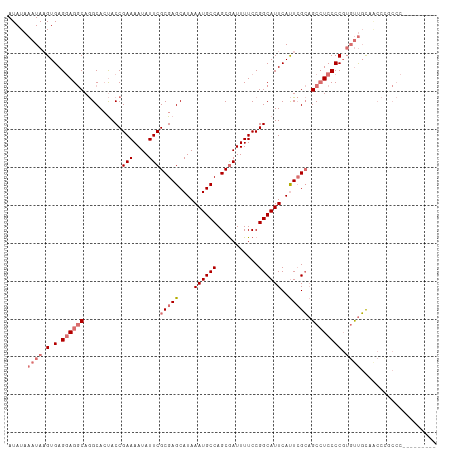
| Location | 17,678,558 – 17,678,662 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Shannon entropy | 0.32993 |
| G+C content | 0.50816 |
| Mean single sequence MFE | -44.73 |
| Consensus MFE | -30.91 |
| Energy contribution | -31.35 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.96 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.78 |
| SVM RNA-class probability | 0.999307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17678558 104 - 24543557 ---------GGGGAGGCGGCAACUCGGGGAGGCUGCGAAUGAAUGCCGGAAAAUCGCUGGCAUUUAUGCUCGCGAAUAUUUUCGGUAGUGCCUGCCUCCUCACUUAUUUAUAU ---------((((((((((((..(((..((...((((((((((((((((.......)))))))))))..)))))....))..)))...)))).))))))))............ ( -49.20, z-score = -5.13, R) >droYak2.chr3L 7824215 95 + 24197627 ------------------AGGGAAAGGGGAGGCGCCUAGUGAAUGCCGGAAAAUCGCUGGCAUUUAUGCUCGCGAAUAUUUUCGGUGGUGCUUGUCUGCUCACUUAUUUAUAU ------------------.(((...(((.(((((((..(((((((((((.......))))))))))).....((((....))))..))))))).))).)))............ ( -34.80, z-score = -2.92, R) >droSec1.super_0 9724892 112 - 21120651 -CUGUUGCGGGGCGGGUUGCAACACGGGGAUGCUGCGAAUGAAUGCCGGAAAAUUGCUGGCAUUUAUGCUCGCGAAUAUUUUCGGUAUUGCCUGCCUCCUCACUUAUUUAUAU -....((.(((((((((....((.((..((((.((((((((((((((((.......)))))))))))..)))))..))))..))))...))))))).)).))........... ( -43.80, z-score = -3.54, R) >droSim1.chr3L 17017591 113 - 22553184 CGGUGGGGGGGGCGGGUUGCAACACGGGGAGGCUGCGAAUGAAUGCCGGAAAAUCGCUGGCAUUUAUGCUCGCGAAUAUUUUCGGUAGUGCCUGCCUCCUCACGUAUUUAUAU ((...((((((((((((.((.((.((..((...((((((((((((((((.......)))))))))))..)))))....))..)))).)))))))))))))).))......... ( -51.10, z-score = -4.26, R) >consensus _________GGGCGGGUUGCAACACGGGGAGGCUGCGAAUGAAUGCCGGAAAAUCGCUGGCAUUUAUGCUCGCGAAUAUUUUCGGUAGUGCCUGCCUCCUCACUUAUUUAUAU .........................((((((((.(((......(((((((((.((((.((((....)))).))))...))))))))).)))..))))))))............ (-30.91 = -31.35 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:22 2011