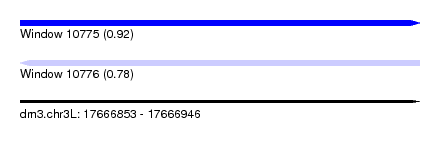
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,666,853 – 17,666,946 |
| Length | 93 |
| Max. P | 0.923963 |

| Location | 17,666,853 – 17,666,946 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 64.51 |
| Shannon entropy | 0.67942 |
| G+C content | 0.59442 |
| Mean single sequence MFE | -38.71 |
| Consensus MFE | -17.89 |
| Energy contribution | -16.98 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
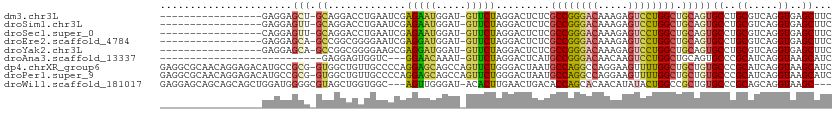
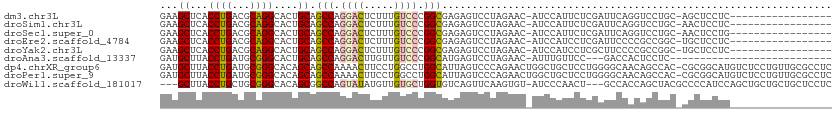
>dm3.chr3L 17666853 93 + 24543557 -----------------GAGGAGCU-GCAGGACCUGAAUCGAGAAUGGAU-GUUCUAGGACUCUCGCCGGGACAAAGAGUCCUGGCUGCAGUGCCUGCGUCAGGUGAGCUUC -----------------..(((((.-(((((..(((....((((.(((..-...)))....))))((((((((.....))))))))..)))..))))).((....))))))) ( -38.40, z-score = -1.89, R) >droSim1.chr3L 17005365 93 + 22553184 -----------------GAGGAGUU-GCAGGACCUGAAUCGAGAAUGGAU-GUUCUAGGACUCUCGCCGGGACAAAGAGUCCUGGCUGCAGUGCCUGCGUCAGGUGAGCUUC -----------------..(((((.-(((((..(((....((((.(((..-...)))....))))((((((((.....))))))))..)))..))))).((....))))))) ( -36.00, z-score = -1.50, R) >droSec1.super_0 9713032 93 + 21120651 -----------------CAGGAGUU-GCAGGACCUGAAUCGAGAAUGGAU-GUUCUAGGACUCUCGCCGGGACAAAGAGUCCUGGCUGCAGUGCCUGCGUCAGGUGAGCUUC -----------------..(((((.-(((((..(((....((((.(((..-...)))....))))((((((((.....))))))))..)))..))))).((....))))))) ( -36.00, z-score = -1.49, R) >droEre2.scaffold_4784 9576371 93 - 25762168 -----------------GAGGAGCA-GCCGGCGGGGAAUCGAGGAUGGAU-GUUCUAGGACUCUCGCCGGGACAAAGAGUCCUGGCUGCAGUGCCUGCGUCAGGUGAGCUUC -----------------((((.(((-((((((((((..((.(((((....-))))).))..)))))))(((((.....)))))))))))...(((((...)))))...)))) ( -41.60, z-score = -2.07, R) >droYak2.chr3L 7810580 93 - 24197627 -----------------GAGGAGCA-GCCGGCGGGGAAGCGAGGAUGGAU-GUUCUAGGACUCUCGCCGGGACAAAGAGUCCUGGCUGCAGUGCCUGCGUCAGGUGAGCUUC -----------------((((.(((-((((((((((.....(((((....-))))).....)))))))(((((.....)))))))))))...(((((...)))))...)))) ( -41.00, z-score = -1.69, R) >droAna3.scaffold_13337 14368894 81 + 23293914 ---------------------------GAGGAGUGGUC---GGAACAAAU-GUUCUAGGACUCAUGCCGGGACAACAAGUCCUGGCUGCAGUGCCCGCAUCAGGUAAGCAUC ---------------------------...((((((..---(((((....-))))).(.(((((.((((((((.....)))))))))).))).))))).))........... ( -29.50, z-score = -1.96, R) >dp4.chrXR_group6 9470331 111 - 13314419 GAGGCGCAACAGGAGACAUGCCGCG-GUGGCUGUUGCCCCAGGAGCAGCCAGUUCUGGGACUAAUGCCAGGCCAGGAAGUUUUGGCUGCUGUGCCCGCAUCAGGUAAGCAUC (.(((((....(....)...((..(-(.(((....))))).))(((((((((((((((..((......)).))))))....)))))))))))))))((.........))... ( -42.20, z-score = 0.27, R) >droPer1.super_9 1500229 111 - 3637205 GAGGCGCAACAGGAGACAUGCCGCG-GUGGCUGUUGCCCCAGGAGCAGCCAGUUCUGGGACUAAUGCCAGGCCAGGAAGUUUUGGCUGCUGUGCCCGCAUCAGGUAAGCAUC (.(((((....(....)...((..(-(.(((....))))).))(((((((((((((((..((......)).))))))....)))))))))))))))((.........))... ( -42.20, z-score = 0.27, R) >droWil1.scaffold_181017 540073 105 - 939937 GAGGAGCAGCAGCAGCUGGAUGGGGCGUAGCUGGUGGC---AGUUGGGAU-ACACUUGAACUGACACCAGCACAACAUAUACUGGCCGCUGUGCCCGCAGCAGGUAAGC--- ..((.(((.((((.((..((((.(..((.(((((((.(---((((.((..-....)).))))).)))))))))..).))).)..)).))))))))).............--- ( -41.50, z-score = -1.28, R) >consensus _________________GAGGAGCG_GCAGGAGCUGAAUCGAGAAUGGAU_GUUCUAGGACUCUCGCCGGGACAAAGAGUCCUGGCUGCAGUGCCUGCGUCAGGUGAGCUUC .........................................(((((.....))))).........((((((((.....)))))))).((..((((.......)))).))... (-17.89 = -16.98 + -0.91)
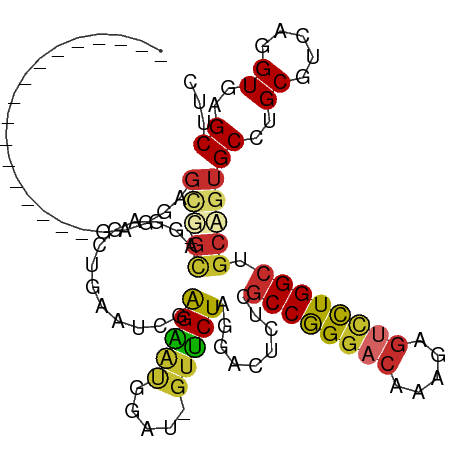
| Location | 17,666,853 – 17,666,946 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 64.51 |
| Shannon entropy | 0.67942 |
| G+C content | 0.59442 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -14.23 |
| Energy contribution | -14.31 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
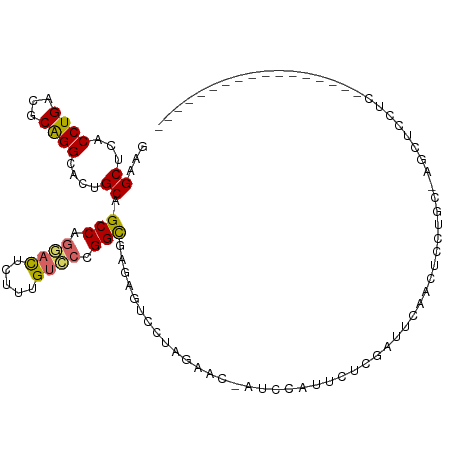
>dm3.chr3L 17666853 93 - 24543557 GAAGCUCACCUGACGCAGGCACUGCAGCCAGGACUCUUUGUCCCGGCGAGAGUCCUAGAAC-AUCCAUUCUCGAUUCAGGUCCUGC-AGCUCCUC----------------- (.(((((....)).(((((.(((...(((.((((.....)))).)))..(((((..((((.-.....)))).))))).))))))))-.))).)..----------------- ( -31.10, z-score = -1.36, R) >droSim1.chr3L 17005365 93 - 22553184 GAAGCUCACCUGACGCAGGCACUGCAGCCAGGACUCUUUGUCCCGGCGAGAGUCCUAGAAC-AUCCAUUCUCGAUUCAGGUCCUGC-AACUCCUC----------------- ...((..((((((.((((...)))).(((.((((.....)))).)))((((((........-....))))))...))))))...))-........----------------- ( -31.00, z-score = -1.91, R) >droSec1.super_0 9713032 93 - 21120651 GAAGCUCACCUGACGCAGGCACUGCAGCCAGGACUCUUUGUCCCGGCGAGAGUCCUAGAAC-AUCCAUUCUCGAUUCAGGUCCUGC-AACUCCUG----------------- ...((..((((((.((((...)))).(((.((((.....)))).)))((((((........-....))))))...))))))...))-........----------------- ( -31.00, z-score = -1.86, R) >droEre2.scaffold_4784 9576371 93 + 25762168 GAAGCUCACCUGACGCAGGCACUGCAGCCAGGACUCUUUGUCCCGGCGAGAGUCCUAGAAC-AUCCAUCCUCGAUUCCCCGCCGGC-UGCUCCUC----------------- ...((((....)).))(((....((((((.((((.....)))).((((.(((((...((..-.....))...)))))..)))))))-))).))).----------------- ( -32.90, z-score = -2.09, R) >droYak2.chr3L 7810580 93 + 24197627 GAAGCUCACCUGACGCAGGCACUGCAGCCAGGACUCUUUGUCCCGGCGAGAGUCCUAGAAC-AUCCAUCCUCGCUUCCCCGCCGGC-UGCUCCUC----------------- ...((((....)).))(((....(((((((((((((((.((....))))))))))).....-..........((......)).)))-))).))).----------------- ( -31.70, z-score = -1.53, R) >droAna3.scaffold_13337 14368894 81 - 23293914 GAUGCUUACCUGAUGCGGGCACUGCAGCCAGGACUUGUUGUCCCGGCAUGAGUCCUAGAAC-AUUUGUUCC---GACCACUCCUC--------------------------- ..(((...((((...))))....)))(((.((((.....)))).)))..((((....((((-....)))).---....))))...--------------------------- ( -23.30, z-score = -0.88, R) >dp4.chrXR_group6 9470331 111 + 13314419 GAUGCUUACCUGAUGCGGGCACAGCAGCCAAAACUUCCUGGCCUGGCAUUAGUCCCAGAACUGGCUGCUCCUGGGGCAACAGCCAC-CGCGGCAUGUCUCCUGUUGCGCCUC ...((.........))((((.(((((((((........))))..(((((..(((((((............)))))))....(((..-...))))))))...))))).)))). ( -37.30, z-score = 0.67, R) >droPer1.super_9 1500229 111 + 3637205 GAUGCUUACCUGAUGCGGGCACAGCAGCCAAAACUUCCUGGCCUGGCAUUAGUCCCAGAACUGGCUGCUCCUGGGGCAACAGCCAC-CGCGGCAUGUCUCCUGUUGCGCCUC ...((.........))((((.(((((((((........))))..(((((..(((((((............)))))))....(((..-...))))))))...))))).)))). ( -37.30, z-score = 0.67, R) >droWil1.scaffold_181017 540073 105 + 939937 ---GCUUACCUGCUGCGGGCACAGCGGCCAGUAUAUGUUGUGCUGGUGUCAGUUCAAGUGU-AUCCCAACU---GCCACCAGCUACGCCCCAUCCAGCUGCUGCUGCUCCUC ---((......))...((((((((((((..(.....((.(((((((((.(((((.......-.....))))---).))))))).)))).....)..))))))).)))))... ( -37.50, z-score = -1.13, R) >consensus GAAGCUCACCUGACGCAGGCACUGCAGCCAGGACUCUUUGUCCCGGCGAGAGUCCUAGAAC_AUCCAUUCUCGAUUCAACUCCUGC_AGCUCCUC_________________ ...((...((((...))))....)).(((.((((.....)))).)))................................................................. (-14.23 = -14.31 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:17 2011