
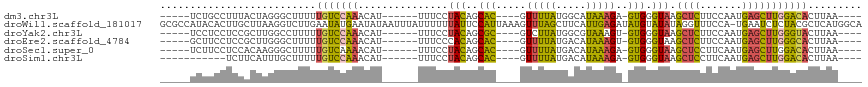
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,663,938 – 17,664,069 |
| Length | 131 |
| Max. P | 0.808263 |

| Location | 17,663,938 – 17,664,035 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.23 |
| Shannon entropy | 0.55241 |
| G+C content | 0.40581 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -11.79 |
| Energy contribution | -11.46 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.702504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17663938 97 + 24543557 -----UCUGCCUUUACUAGGGCUUUUUGUCCAAACAU------UUUCCUACAGCAC----GUUUUAUGGCAUAAAGA-GUGGGUAAGCUCUUCCAAUGAGCUUGGACACUUAA---- -----..((((.......((((.....))))((((..------.............----))))...))))....((-(((..(((((((.......)))))))..)))))..---- ( -26.46, z-score = -1.80, R) >droWil1.scaffold_181017 535531 116 - 939937 GCGCCAUACACUUGCUUAAGGUCUUGAAUAUGAAUAUAAUUUAUUUUUUAUUCCAUUAAAGUUUAGCUUCAUUGAGAUAUGUAUAUAGGUUUCCA-UGAAUCUCUACGCUCAUGGCA ..(((((..((((.....))))...(((((.(((((.....)))))..))))).......((..((.(((((.(((((..........))))).)-)))).))..))....))))). ( -20.60, z-score = -0.37, R) >droYak2.chr3L 7807870 97 - 24197627 -----UCCUCCUCCGCUUGGCCUUUUUGUCCAAACAU------UUUCCUACAGCGC----GUCUUAUGGCGUAAAGU-GUGGGUAAGCUCUUCCAAUGAGCUUGGGUACUUAA---- -----.......(((.((((.(.....).)))).)..------...((((((..((----(((....)))))....)-)))))(((((((.......))))))))).......---- ( -24.70, z-score = -0.89, R) >droEre2.scaffold_4784 9573524 97 - 25762168 -----GCUUCCUCCGCUUGGGCUUUUUGUCCAAACAU------UUUCCCACAGCAC----GUUUUAUGACAUAAAGU-GUGGGUAAGCUCUUCCAAUGAGCUUGGGCACUUAA---- -----(((......(.((((((.....)))))).)..------...((((((....----(((....)))......)-)))))(((((((.......))))))))))......---- ( -26.50, z-score = -1.43, R) >droSec1.super_0 9710081 97 + 21120651 -----UCUUCCUCCACAAGGGCUUUUUGUCAAAACAU------UUUCCUACAGCAC----GUUUUAUGACAUAAAGA-GUGGGUAAGCUCCUUCAAUGAGCUUGGACACUUAA---- -----...((((.....))))((((.(((((((((..------.............----))))..))))).))))(-(((..(((((((.......)))))))..))))...---- ( -22.76, z-score = -1.39, R) >droSim1.chr3L 17002400 91 + 22553184 -----------UCUUCAUUUGCUUUUUGUCCAAACAU------UUUCCUACAGCAC----GUUUUAUGACAUAAAGA-GUGGGUAAGCUCCUUCAAUGAGCUUGGACACUUAA---- -----------..........((((.((((.((((..------.............----))))...)))).))))(-(((..(((((((.......)))))))..))))...---- ( -19.36, z-score = -1.42, R) >consensus _____UCUUCCUCCACUAGGGCUUUUUGUCCAAACAU______UUUCCUACAGCAC____GUUUUAUGACAUAAAGA_GUGGGUAAGCUCUUCCAAUGAGCUUGGACACUUAA____ ..........................(((((...............(((((.........(((....)))........))))).((((((.......)))))))))))......... (-11.79 = -11.46 + -0.32)


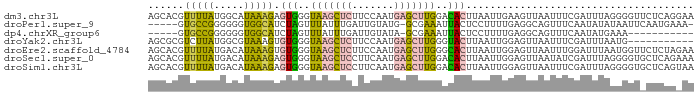
| Location | 17,663,978 – 17,664,069 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 62.58 |
| Shannon entropy | 0.70574 |
| G+C content | 0.39419 |
| Mean single sequence MFE | -20.66 |
| Consensus MFE | -9.06 |
| Energy contribution | -7.83 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.808263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17663978 91 + 24543557 AGCACGUUUUAUGGCAUAAAGAGUGGGUAAGCUCUUCCAAUGAGCUUGGACACUUAAUUGAAGUUAAUUUCGAUUUAGGGGUUCUCAGGAA .((.(((...))))).....(((((..(((((((.......)))))))..))))).......(..(((..(......)..)))..)..... ( -21.90, z-score = -0.96, R) >droPer1.super_9 1497620 84 - 3637205 -----GUGCCGGGGGGUGGCAUCUAGUUUAUUUGAUUGUAUG-GCGAAAUUACUCCUUUUGAGGCAGUUUCAAUAUAUAAUUCAAUGAAA- -----((((((.....))))))....((((((.(((((((((-..((((((.(((.....)))..))))))..))))))))).)))))).- ( -22.40, z-score = -1.88, R) >dp4.chrXR_group6 9467688 74 - 13314419 -----GUGCCGGGGGGUGGCAUCUAGUUUAUUUGAUUGUAUA-GCGAAAUUACUCCUUUUGAGGCAGUUUCAAUAUGAAA----------- -----.((((((((((((((((.(((((.....))))).)).-)).....))))))))....)))).((((.....))))----------- ( -14.10, z-score = 0.51, R) >droYak2.chr3L 7807910 80 - 24197627 AGCGCGUCUUAUGGCGUAAAGUGUGGGUAAGCUCUUCCAAUGAGCUUGGGUACUUAAUUGGAGUUAAUUUCGAUUUAAUG----------- ...(((((....))))).....(((..(((((((.......)))))))..)))..((((((((....)))))))).....----------- ( -19.40, z-score = -1.06, R) >droEre2.scaffold_4784 9573564 91 - 25762168 AGCACGUUUUAUGACAUAAAGUGUGGGUAAGCUCUUCCAAUGAGCUUGGGCACUUAAUUGGAGUUAAUUUGGAUUUAAUGGUUCUCUAGAA .....(((....)))...((((((...(((((((.......))))))).))))))..((((((...(((.......)))....)))))).. ( -19.00, z-score = -0.08, R) >droSec1.super_0 9710121 91 + 21120651 AGCACGUUUUAUGACAUAAAGAGUGGGUAAGCUCCUUCAAUGAGCUUGGACACUUAAUUGGAGUUAAUAUCGAUUUAGGGGUGCUCAGAAA ((((((((....))).....(((((..(((((((.......)))))))..))))).........................)))))...... ( -22.80, z-score = -1.41, R) >droSim1.chr3L 17002434 91 + 22553184 AGCACGUUUUAUGACAUAAAGAGUGGGUAAGCUCCUUCAAUGAGCUUGGACACUUAAUUGGAGUUAAUUUCGAUUUAGGGGUGCUCAGUAA ((((((((....))).....(((((..(((((((.......)))))))..)))))((((((((....)))))))).....)))))...... ( -25.00, z-score = -2.03, R) >consensus AGCACGUUUUAUGGCAUAAAGUGUGGGUAAGCUCUUUCAAUGAGCUUGGACACUUAAUUGGAGUUAAUUUCGAUUUAAGGGU_CUCAG_AA ......(((((.....))))).(((..(((((((.......)))))))..)))...................................... ( -9.06 = -7.83 + -1.24)
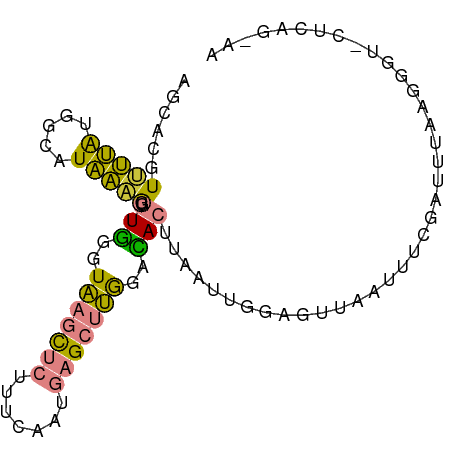

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:15 2011