
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,636,581 – 17,636,682 |
| Length | 101 |
| Max. P | 0.780260 |

| Location | 17,636,581 – 17,636,682 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 88.39 |
| Shannon entropy | 0.24301 |
| G+C content | 0.60256 |
| Mean single sequence MFE | -40.68 |
| Consensus MFE | -34.62 |
| Energy contribution | -34.33 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.780260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
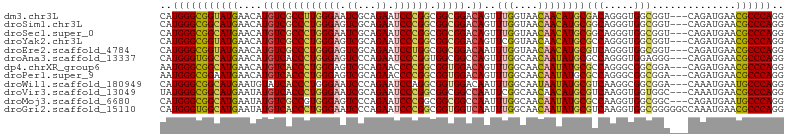
>dm3.chr3L 17636581 101 - 24543557 CAUGGGCGGUAUGAACAUGUCGCCUUGGGAAUCGCAGAAUCCCGGCGGCGGACAGUUUGGUAACAACAUGCGACAGGGUGGCGGU---CAGAUGAACGCCCAGG ..((((((...(((...((((((((((....(((((.....(((....)))...(((....)))....))))))))))))))).)---))......)))))).. ( -38.00, z-score = -1.53, R) >droSim1.chr3L 16976904 101 - 22553184 CAUGGGCGGCAUGAACAUGUCGCCCUGGGAGUCGCAGAAUCCCGGCGGCGGACAGUUUGGUAACAACAUGCGGCAGGGUGGCGGU---CAGAUGAACGCCCAGG ..((((((...(((...(((((((((((((.((...)).)))).((.(((....(((....)))....))).))))))))))).)---))......)))))).. ( -42.60, z-score = -1.69, R) >droSec1.super_0 9682674 101 - 21120651 CAUGGGCGGCAUGAACAUGUCGCCCUGGGAAUCGCAGAAUCCCGGCGGCGGACAGUUUGGUAACAACAUGCGGCAGGGUGGCGGU---CAGAUGAACGCCCAGG ..((((((...(((...(((((((((((((.((...)).)))).((.(((....(((....)))....))).))))))))))).)---))......)))))).. ( -42.60, z-score = -1.92, R) >droYak2.chr3L 7778512 101 + 24197627 CAUGGGCGGUAUGAACAUGUCGCCCUGGGAGUCGCAGAAUCCCGGCGGCGGACAGUUCGGUAACAACAUGCGCCAGGGUGGCGGU---CAGAUGAACGCCCAGG ..((((((((.(((((.(((((((((((((.((...)).)))))).))).)))))))))...))......((((.....))))..---........)))))).. ( -44.20, z-score = -2.06, R) >droEre2.scaffold_4784 9544180 101 + 25762168 CAUGGGCGGUAUGAACAUGUCGCCCUGGGAGUCGCAGAAUCCUGGCGGCGGACAAUUUGGUAACAACAUGCGUCAGGGUGGCGGU---CAGAUGAACGCCCAGG ..((((((...(((...(((((((((((..(((((((....)).)))))..........(((......))).))))))))))).)---))......)))))).. ( -40.90, z-score = -2.06, R) >droAna3.scaffold_13337 14340263 101 - 23293914 CAUGGGUGGCAUGAACAUGUCACCCUGGGAGUCCCAGAAUCCCGGUGGCGGCCAGUUUGGCAACAAUAUGCGCCAGGGUGGAGGG---CAGAUGAACGCCCAGG ...(((((((((....)))))))))((((...))))...((((..(((((..((..(((....)))..)))))))..).)))(((---(........))))... ( -41.40, z-score = -0.96, R) >dp4.chrXR_group6 9437813 101 + 13314419 AAUGGGCGGCAUGAACAUGUCACCCUGGGAGUCGCAGAACCCCGGCGGUGGACAGUUUGGCAACAAUAUGCGCCAGGGCGGCGGA---CAGAUGAACGCCCAGG ..((((((((((((((.((((((((((((.((......))))))).))).)))))))((....)).)))))(((.....)))...---........)))))).. ( -42.10, z-score = -1.72, R) >droPer1.super_9 1469090 101 + 3637205 AAUGGGCGGAAUGAACAUGUCACCCUGGGAGUCGCAGAACCCCGGCGGUGGACAGUUUGGCAACAAUAUGCGCCAGGGCGGCGGA---CAGAUGAACGCCCAGG ..((((((.........((((((((((((.((......))))))).))).))))((((((((......)))(((.....)))...---)))))...)))))).. ( -39.30, z-score = -1.31, R) >droWil1.scaffold_180949 1478017 101 - 6375548 CAUGGGCGGCAUGAAUGUAUCACCCUGGGAAUCCCAGAAUCCAGGCGGUGGACAAUUUGGCAAUAAUAUGCGUCAAGGCGGCGGA---CAAAUGAAUGCCCAGG ..((((((.......(((.(((((((.(((.((...)).))).)).))))))))((((((((......)))(((.....)))...---)))))...)))))).. ( -29.50, z-score = 0.42, R) >droVir3.scaffold_13049 18438934 101 + 25233164 UAUGGGCGGCAUGAAUAUGUCACCCUGGGAAUCGCAGAAUCCCGGCGGCGGCCAAUUCGGCAACAACAUGCGUCAAGGUGGUGGC---CAAAUGAACGCCCAGG ..(((((((((((.....(((.((((((((.((...)).)))))).)).)))......(....)..))))).....(((....))---).......)))))).. ( -38.90, z-score = -1.17, R) >droMoj3.scaffold_6680 19637853 101 + 24764193 CAUGGGCGGCAUGAAUAUGUCGCCGUGGGAGUCCCAGAAUCCCGGCGGCGGCCAAUUUGGCAACAAUAUGCGCCAAGGUGGCGGC---CAGAUGAAUGCCCAGG ..((((((((((....)))))(((..((((.((...)).)))))))(((.((((.((((((..........)))))).)))).))---)........))))).. ( -45.60, z-score = -1.92, R) >droGri2.scaffold_15110 6001132 104 - 24565398 CAUGGGUGGCAUGAAUAUGUCACCCUGGGAAUCCCAGAAUCCCGGCGGUGGUCAAUUUGGCAACAAUAUGCGUCAAGGUGGCGGGGGCCAAAUGAACGCCCAGG ..(((((((((((....(((((((((((((.((...)).)))))).))))).))..(((....)))))))).(((...((((....))))..))).)))))).. ( -43.00, z-score = -1.85, R) >consensus CAUGGGCGGCAUGAACAUGUCACCCUGGGAGUCGCAGAAUCCCGGCGGCGGACAGUUUGGCAACAACAUGCGCCAGGGUGGCGGU___CAGAUGAACGCCCAGG ..(((((((((((....(((((((((((((.((...)).)))))).))))).))..(((....))))))))(((.....)))..............)))))).. (-34.62 = -34.33 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:12 2011