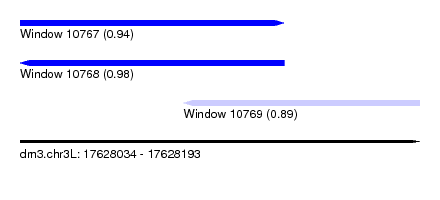
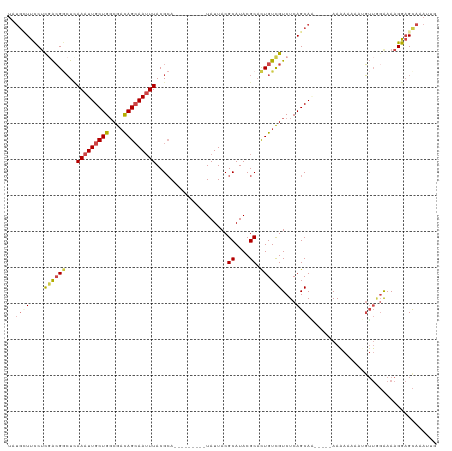
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,628,034 – 17,628,193 |
| Length | 159 |
| Max. P | 0.975874 |

| Location | 17,628,034 – 17,628,139 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.73 |
| Shannon entropy | 0.40582 |
| G+C content | 0.42130 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -18.19 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
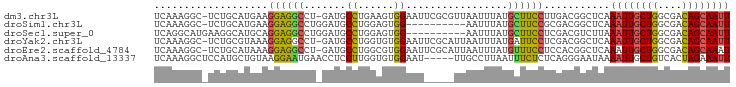
>dm3.chr3L 17628034 105 + 24543557 CGAUUUGCUCCUUUCCCAACAUUUUUUUU-----UUGCUGAGACGACAGUGCGUAUGCGUAAUA---------UGCGUAAAUUGCUGUCGCCAGCAAUUUGAGCCGUCAAGGAAGCAUA .....((((((..................-----((((((.(.((((((((..(((((((...)---------))))))...))))))))))))))).((((....)))))).)))).. ( -32.00, z-score = -2.64, R) >droSim1.chr3L 16967301 110 + 22553184 CCAUUUGCUCCUUUUCCAACAUUUUUUUUCUUUUUUGCUGAGACGACAGUGCGUAUGCGUAAUA---------UGCGUAAAUUGCUGUCGCCAGCAAUUUGAGCCGUCGCGGAAGCAUA .....((((((.................((....((((((.(.((((((((..(((((((...)---------))))))...)))))))))))))))...))((....)))).)))).. ( -32.80, z-score = -2.62, R) >droSec1.super_0 9674267 109 + 21120651 CAAUUUGCUCCUUUUCCAACAUUUUUUUU-UUUUUUGCUGAGACGACAGUGCGUAUGCGUAAUA---------UGCGUAAAUUGCUGUCGCCAGCAAUUUAAGACGUCGAGGAAGCAUA .....((((((((................-....((((((.(.((((((((..(((((((...)---------))))))...))))))))))))))).....(....))))).)))).. ( -31.80, z-score = -2.81, R) >droYak2.chr3L 7769991 110 - 24197627 CAGUUUGCUCCUUUUCGAAC----AUUUU-----GUGCUGAGACGACAGUGCGUAUGCGUAAUACGUGUAAUAUGCGUAAAUUGCUGUCGCCAGCAAUUUGAGCCGUCGAGGAAUCAUA ........(((((..((..(----(....-----.(((((.(.((((((((..((((((((.((....)).))))))))...))))))))))))))...))...))..)))))...... ( -34.40, z-score = -1.96, R) >droEre2.scaffold_4784 9535689 99 - 25762168 --GUAUACUCCUUUCGUAAC----UUUUU-----CUGCUGAGACGACAGUGCGUAUGCGUAAUA---------UGCGUAAUUUGCUGUCGCCAGCAAUUUGAGCCGUGGAGGAAACAUA --((...((((...((...(----((...-----.(((((.(.((((((((..(((((((...)---------))))))...))))))))))))))....))).)).))))...))... ( -32.60, z-score = -2.84, R) >droAna3.scaffold_13337 14331834 103 + 23293914 CAAUUUGCGCCUUUUCUUUACUCCUUAUU--UUUAUGCUGAGACAACAAGGCGUAUGCGUAAUA---------UCCGUAAAUUUCUAGUGACAGCAAUUUUAUUCCCUGAGAGA----- .....((((((((.......(((..(((.--...)))..))).....))))))))((((.....---------..))))..((((.(((((.(....).)))))....))))..----- ( -14.30, z-score = 0.76, R) >consensus CAAUUUGCUCCUUUUCCAACAUUUUUUUU_____UUGCUGAGACGACAGUGCGUAUGCGUAAUA_________UGCGUAAAUUGCUGUCGCCAGCAAUUUGAGCCGUCGAGGAAGCAUA ...................................(((((.(.((((((((..((((((..............))))))...))))))))))))))....................... (-18.19 = -18.47 + 0.28)

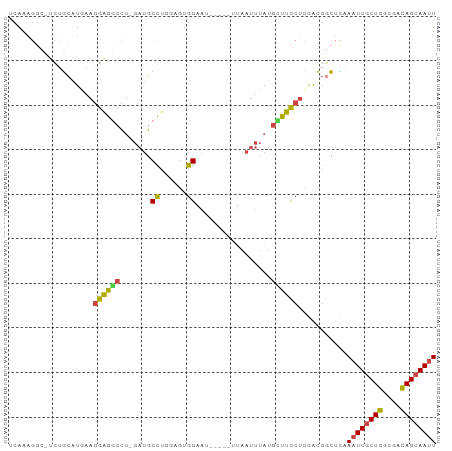
| Location | 17,628,034 – 17,628,139 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.73 |
| Shannon entropy | 0.40582 |
| G+C content | 0.42130 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -17.77 |
| Energy contribution | -18.33 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.975874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17628034 105 - 24543557 UAUGCUUCCUUGACGGCUCAAAUUGCUGGCGACAGCAAUUUACGCA---------UAUUACGCAUACGCACUGUCGUCUCAGCAA-----AAAAAAAAUGUUGGGAAAGGAGCAAAUCG ..((((.(((((((((...(((((((((....))))))))).....---------......((....)).))))).((((((((.-----........))))))))))))))))..... ( -36.40, z-score = -4.59, R) >droSim1.chr3L 16967301 110 - 22553184 UAUGCUUCCGCGACGGCUCAAAUUGCUGGCGACAGCAAUUUACGCA---------UAUUACGCAUACGCACUGUCGUCUCAGCAAAAAAGAAAAAAAAUGUUGGAAAAGGAGCAAAUGG ..((((((.(((((((...(((((((((....))))))))).....---------......((....)).))))))).((((((..............))))))....))))))..... ( -32.84, z-score = -2.76, R) >droSec1.super_0 9674267 109 - 21120651 UAUGCUUCCUCGACGUCUUAAAUUGCUGGCGACAGCAAUUUACGCA---------UAUUACGCAUACGCACUGUCGUCUCAGCAAAAAA-AAAAAAAAUGUUGGAAAAGGAGCAAAUUG ..((((.(((((((((..((((((((((....)))))))))).)).---------......((....))...))))((.(((((.....-........)))))))..)))))))..... ( -30.72, z-score = -3.44, R) >droYak2.chr3L 7769991 110 + 24197627 UAUGAUUCCUCGACGGCUCAAAUUGCUGGCGACAGCAAUUUACGCAUAUUACACGUAUUACGCAUACGCACUGUCGUCUCAGCAC-----AAAAU----GUUCGAAAAGGAGCAAACUG ..((.(((((.((((((..(((((((((....)))))))))............(((((.....)))))....))))))((((((.-----....)----))).))..)))))))..... ( -29.70, z-score = -2.27, R) >droEre2.scaffold_4784 9535689 99 + 25762168 UAUGUUUCCUCCACGGCUCAAAUUGCUGGCGACAGCAAAUUACGCA---------UAUUACGCAUACGCACUGUCGUCUCAGCAG-----AAAAA----GUUACGAAAGGAGUAUAC-- ...............((((...((((((((((((((.......)).---------......((....))..))))))..))))))-----.....----....(....)))))....-- ( -23.30, z-score = -1.06, R) >droAna3.scaffold_13337 14331834 103 - 23293914 -----UCUCUCAGGGAAUAAAAUUGCUGUCACUAGAAAUUUACGGA---------UAUUACGCAUACGCCUUGUUGUCUCAGCAUAAA--AAUAAGGAGUAAAGAAAAGGCGCAAAUUG -----(((.....)))......((((.(((............((..---------.....))..(((.(((((((.............--))))))).))).......))))))).... ( -16.02, z-score = 0.58, R) >consensus UAUGCUUCCUCGACGGCUCAAAUUGCUGGCGACAGCAAUUUACGCA_________UAUUACGCAUACGCACUGUCGUCUCAGCAA_____AAAAAAAAUGUUGGAAAAGGAGCAAAUUG ..((((....((((((...(((((((((....)))))))))....................((....)).))))))....))))................................... (-17.77 = -18.33 + 0.56)

| Location | 17,628,099 – 17,628,193 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 72.36 |
| Shannon entropy | 0.52146 |
| G+C content | 0.47880 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -13.35 |
| Energy contribution | -12.80 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
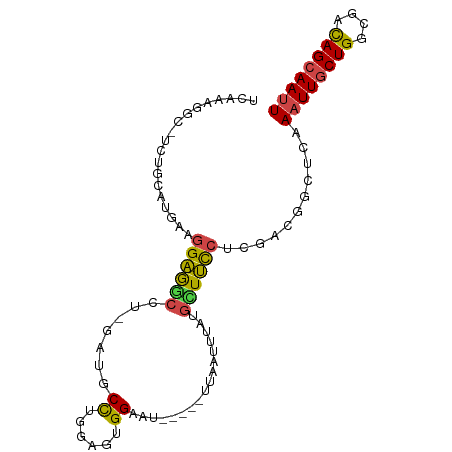
>dm3.chr3L 17628099 94 - 24543557 UCAAAGGC-UCUGCAUGAAGGAGGCCU-GAUGCCUGAAGUGGAAUUCGCGUUAAUUUAUGCUUCCUUGACGGCUCAAAUUGCUGGCGACAGCAAUU ....((((-.((.(.....).))))))-((.(((......((((...((((......)))))))).....))))).((((((((....)))))))) ( -29.70, z-score = -1.23, R) >droSim1.chr3L 16967371 85 - 22553184 UCAAAGGC-UCUGCAUGAAGGAGGCCUGGAUGCCUGGAGUGG----------AAUUUAUGCUUCCGCGACGGCUCAAAUUGCUGGCGACAGCAAUU ....((((-.((.(.....).)))))).((.(((.(..((((----------((.......))))))..)))))).((((((((....)))))))) ( -32.80, z-score = -2.20, R) >droSec1.super_0 9674336 86 - 21120651 UCAGGCAUGAAGGCAUGCAGGAGGCCUGGAUGCCUGGAGUGG----------AAUUUAUGCUUCCUCGACGUCUUAAAUUGCUGGCGACAGCAAUU ..((((....((((((.((((...)))).))))))(..(.((----------((.......)))).)..)))))..((((((((....)))))))) ( -34.10, z-score = -2.73, R) >droYak2.chr3L 7770061 94 + 24197627 UCAAAGGC-UCUGCGUAAAGGAGGCCU-GAUGCCUGGUGUGGAAUUCGCAUUAAUUUAUGAUUCCUCGACGGCUCAAAUUGCUGGCGACAGCAAUU ....((((-.((.(.....).))))))-((.(((...((.(((((...(((......)))))))).))..))))).((((((((....)))))))) ( -30.70, z-score = -1.75, R) >droEre2.scaffold_4784 9535748 94 + 25762168 UCAAAGGC-UCUGCAUAAAGGAGGCCU-GAUGCCUGGCGUGGAAUUCGCAUUAAUUUAUGUUUCCUCCACGGCUCAAAUUGCUGGCGACAGCAAAU ....((((-.((.(.....).))))))-((.((((((...((((...((((......)))))))).))).)))))...((((((....)))))).. ( -31.60, z-score = -1.98, R) >droAna3.scaffold_13337 14331902 91 - 23293914 UCAAAGGCUCCAUGCUGUAAGGAAUGAACCUCCUUGGUGUGGAAU-----UUGCCUUAAUUUCUCUCAGGGAAUAAAAUUGCUGUCACUAGAAAUU ...(((((((((((((..(((((.......)))))))))))))..-----..)))))(((((((..(((.((......)).))).....))))))) ( -24.80, z-score = -2.01, R) >consensus UCAAAGGC_UCUGCAUGAAGGAGGCCU_GAUGCCUGGAGUGGAAU_____UUAAUUUAUGCUUCCUCGACGGCUCAAAUUGCUGGCGACAGCAAUU ...................(((((((.....)((......)).................))))))...........((((((((....)))))))) (-13.35 = -12.80 + -0.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:11 2011