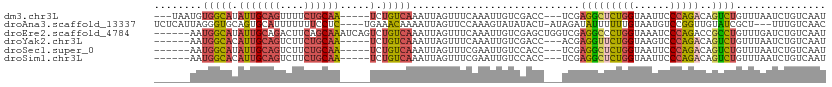
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,624,551 – 17,624,652 |
| Length | 101 |
| Max. P | 0.973199 |

| Location | 17,624,551 – 17,624,652 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.53 |
| Shannon entropy | 0.47933 |
| G+C content | 0.39853 |
| Mean single sequence MFE | -23.72 |
| Consensus MFE | -10.44 |
| Energy contribution | -9.58 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
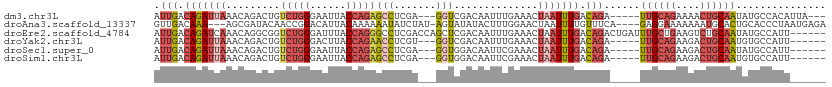
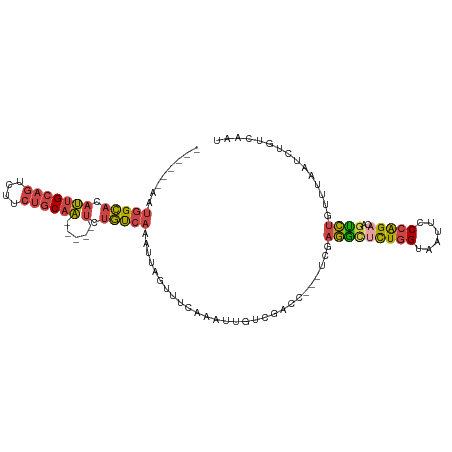
>dm3.chr3L 17624551 101 + 24543557 AUUGACAGAUUAAACAGACUGUCUGGGAAUUACCAGAGCCUCGA---GGUCGACAAUUUGAAACUAAUUUGACAGA-----UUGCAGAAAACUGCAAUAUGCCACAUUA--- .(((.(((((((..((((.(((((((......)))((.((....---)))))))).))))....))))))).)))(-----((((((....)))))))...........--- ( -24.70, z-score = -2.88, R) >droAna3.scaffold_13337 14328647 104 + 23293914 GUUGACAAA---AGCGAUACAACCGGACAUUACAAAAAAUAUCUAU-AGUAUAUACUUUGGAACUAAUUUUGUUUCA----GAGGAAAAAAAUGCACUGCACCCUAAUGAGA .........---.(((((((....(((.(((......))).)))..-.))))...(((((((((.......))))))----)))........)))................. ( -14.30, z-score = -0.35, R) >droEre2.scaffold_4784 9532378 106 - 25762168 AUUGACAGAUCAAACAGGCGGUCUGGGAUUUACCAGGGCCUCGACCAGCUCGACAAUUUGAAACUAAUUUGACAGACUGAUUUGCUGAAGUCUGCAAUAUGCCAUU------ .((((....))))...((((((((((......))).))))((((.....))))...............(((.((((((..........)))))))))...)))...------ ( -25.90, z-score = -0.79, R) >droYak2.chr3L 7766559 98 - 24197627 AUUGACAGAUUAAACAGACUGUCUGGGACUUACCAGAACCUCGU---GGUCGACAAUUUGAAACUAAUUUGACAGA-----UUGCAGAAGACUGCAAUGUGCCAUU------ .....(((((((..((((.(((((((......)))((.((....---)))))))).))))....))))))).((.(-----((((((....))))))).)).....------ ( -26.90, z-score = -2.85, R) >droSec1.super_0 9670929 98 + 21120651 AUUGACAGAUUAAACAGACUGUCUGGGAAUUACCAGAGCCUCGA---GGUGGACAAUUCGAAACUAAUUUGACAGA-----UUGCAGAAGACUGCAAUAUGCCAUU------ .(((.(((((((.........(((((......)))))...((((---(........)))))...))))))).)))(-----((((((....)))))))........------ ( -24.30, z-score = -2.30, R) >droSim1.chr3L 16963880 98 + 22553184 AUUGACAGAUUAAACAGACUGUCUGGGAAUUACCAGAGCCUCGA---GGUGGACAAUUCGAAACUAAUUUGACAGA-----UUGCAGAAGACUGCAAUGUGCCAUU------ .....(((((((.........(((((......)))))...((((---(........)))))...))))))).((.(-----((((((....))))))).)).....------ ( -26.20, z-score = -2.41, R) >consensus AUUGACAGAUUAAACAGACUGUCUGGGAAUUACCAGAGCCUCGA___GGUCGACAAUUUGAAACUAAUUUGACAGA_____UUGCAGAAGACUGCAAUAUGCCAUU______ ((((.(((........((...(((((......)))))...)).........................((((.(((......)))))))...))))))).............. (-10.44 = -9.58 + -0.85)
| Location | 17,624,551 – 17,624,652 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.53 |
| Shannon entropy | 0.47933 |
| G+C content | 0.39853 |
| Mean single sequence MFE | -25.15 |
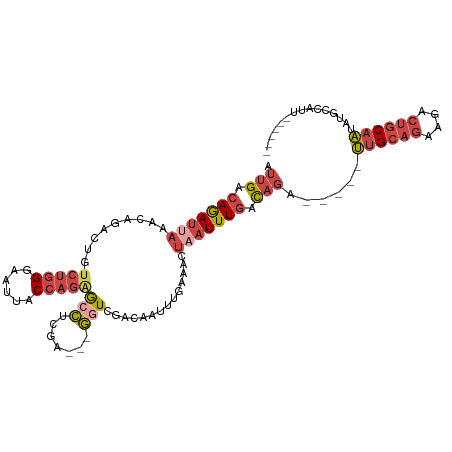
| Consensus MFE | -12.26 |
| Energy contribution | -12.32 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.973199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
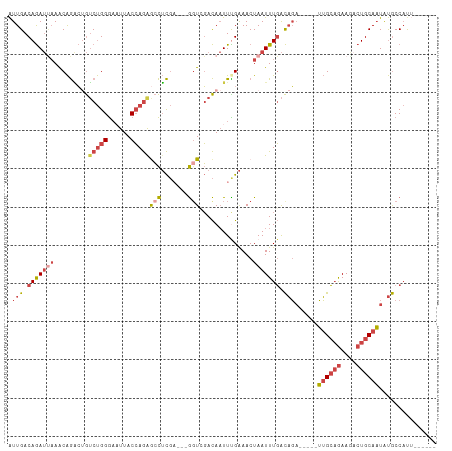
>dm3.chr3L 17624551 101 - 24543557 ---UAAUGUGGCAUAUUGCAGUUUUCUGCAA-----UCUGUCAAAUUAGUUUCAAAUUGUCGACC---UCGAGGCUCUGGUAAUUCCCAGACAGUCUGUUUAAUCUGUCAAU ---((((.(((((.(((((((....))))))-----).))))).))))....((.((((((((((---....)).))(((......))))))))).)).............. ( -26.20, z-score = -2.94, R) >droAna3.scaffold_13337 14328647 104 - 23293914 UCUCAUUAGGGUGCAGUGCAUUUUUUUCCUC----UGAAACAAAAUUAGUUCCAAAGUAUAUACU-AUAGAUAUUUUUUGUAAUGUCCGGUUGUAUCGCU---UUUGUCAAC ...((..(((((((((...(((((((((...----.)))).))))).....((..(((....)))-...((((((......)))))).))))))))).))---..))..... ( -12.90, z-score = 0.90, R) >droEre2.scaffold_4784 9532378 106 + 25762168 ------AAUGGCAUAUUGCAGACUUCAGCAAAUCAGUCUGUCAAAUUAGUUUCAAAUUGUCGAGCUGGUCGAGGCCCUGGUAAAUCCCAGACCGCCUGUUUGAUCUGUCAAU ------..(((((..(((((((((..........)))))).)))..............(((((((.(((...((..((((......)))).))))).))))))).))))).. ( -27.90, z-score = -1.40, R) >droYak2.chr3L 7766559 98 + 24197627 ------AAUGGCACAUUGCAGUCUUCUGCAA-----UCUGUCAAAUUAGUUUCAAAUUGUCGACC---ACGAGGUUCUGGUAAGUCCCAGACAGUCUGUUUAAUCUGUCAAU ------..(((((.(((((((....))))))-----).))))).(((((...((.((((((((((---....)))).(((......))))))))).)).)))))........ ( -26.90, z-score = -2.97, R) >droSec1.super_0 9670929 98 - 21120651 ------AAUGGCAUAUUGCAGUCUUCUGCAA-----UCUGUCAAAUUAGUUUCGAAUUGUCCACC---UCGAGGCUCUGGUAAUUCCCAGACAGUCUGUUUAAUCUGUCAAU ------..(((((.(((((((....))))))-----).)))))....((((((((..........---)))))))).(((......)))(((((..........)))))... ( -28.50, z-score = -4.04, R) >droSim1.chr3L 16963880 98 - 22553184 ------AAUGGCACAUUGCAGUCUUCUGCAA-----UCUGUCAAAUUAGUUUCGAAUUGUCCACC---UCGAGGCUCUGGUAAUUCCCAGACAGUCUGUUUAAUCUGUCAAU ------..(((((.(((((((....))))))-----).)))))....((((((((..........---)))))))).(((......)))(((((..........)))))... ( -28.50, z-score = -3.76, R) >consensus ______AAUGGCACAUUGCAGUCUUCUGCAA_____UCUGUCAAAUUAGUUUCAAAUUGUCGACC___UCGAGGCUCUGGUAAUUCCCAGACAGUCUGUUUAAUCUGUCAAU ........(((((..((((((....))))))........................................(((((((((......)))))..))))........))))).. (-12.26 = -12.32 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:08 2011