| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,619,439 – 17,619,549 |
| Length | 110 |
| Max. P | 0.865942 |

| Location | 17,619,439 – 17,619,549 |
|---|---|
| Length | 110 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 67.72 |
| Shannon entropy | 0.64725 |
| G+C content | 0.43410 |
| Mean single sequence MFE | -24.11 |
| Consensus MFE | -12.02 |
| Energy contribution | -10.92 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.865942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17619439 110 + 24543557 GGCCUGAUGGCAAUAUUCUGCACCCAGAGCAAUGACUUCGCUAUCGGCUAUCCCAUAGGUGAGUUCAUGUUACAAGAACCUGGAUAUUGCAUCUAGAAAUAUAUGCAACU ((((.((((((....(((((....)))))....(....)))))))))))((((...((((..((.......))....)))))))).((((((.((....)).)))))).. ( -28.40, z-score = -0.88, R) >droSim1.chr3L 16958769 99 + 22553184 GGUCUGAUGGCUAUAUUCUGCACCCAGAGCAAUGACUUUGCCAUCGGAUAUCCCAUAGGUGAGUUCAGGUCAUAAGAACCUGAAUUUUGCAUUCAAAAC----------- .((((((((((.......(((.......)))........)))))))))).........(..((((((((((....).))))))).))..).........----------- ( -29.16, z-score = -2.16, R) >droSec1.super_0 9665810 99 + 21120651 GGCCUGAUGGCUAUAUUCUGCACCCAGAGCAAUGACUUCGCUAUCGGAUAUCCCAUAGGUGAGUUCAGGUCAUAAGAACCUGAAUUUUGCAUUCAAAAC----------- ((((....))))....((((....))))((((.....(((((((.((....)).))).))))(((((((((....).)))))))).)))).........----------- ( -26.60, z-score = -1.45, R) >droYak2.chr3L 7761446 89 - 24197627 GGCCUGAUGGCUAUAUUCUGCACCCAGAGCAAUGACUUCGCUAUCGGCUAUCCCAUAGGCAAGU----------GUAACCUGUAUACUUUAUCCCAAAU----------- ((((.((((((....(((((....)))))....(....)))))))))))........((.((((----------(((.....)))))))....))....----------- ( -20.10, z-score = -0.48, R) >droEre2.scaffold_4784 9527174 99 - 25762168 GGCUUGAUGGCUAUAUUCUGCACCCAGAGCAAUGACUUCGCUAUCGGCUAUCCCAUAGGUGAGUUGAUGCCGGAGAAACCUCGAUACUUUAUCCAAAAU----------- .((((....((........)).....)))).....(((((.((((((((..((....))..)))))))).)))))........................----------- ( -22.50, z-score = 0.02, R) >droAna3.scaffold_13337 14321596 95 + 23293914 GGCUUGAUGGCAAUUUUCUGCACUCAGAGUAACGACUUUGCCAUCGGCUAUCCCAUAGGUUAGUU--UAAAAUUGUUAAAAAAAAUGUGUAAAAAAU------------- ((((.((((((((..(((((....)))))........))))))))))))....((((......((--(((.....))))).....))))........------------- ( -19.10, z-score = -0.94, R) >dp4.chrXR_group6 9417188 91 - 13314419 GGUUUGAUGGCCAUUUUCUGUACGCAGAGCAAUGAUUUUGCCAUCGGCUAUCCCAUAGGUGGGUU----UCAUCAUCAUCUGUUCAAUGCAUCUU--------------- ((...(((((((....((((....))))((((.....))))....)))))))))((((((((...----......))))))))............--------------- ( -24.50, z-score = -1.05, R) >droPer1.super_9 1448335 91 - 3637205 GGUUUGAUGGCCAUUUUCUGUACGCAGAGCAAUGAUUUUGCCAUCGGCUAUCCCAUAGGUGGGUU----UCAUCAUCAUCUGUUCAAUGCAUCUU--------------- ((...(((((((....((((....))))((((.....))))....)))))))))((((((((...----......))))))))............--------------- ( -24.50, z-score = -1.05, R) >droWil1.scaffold_180949 1448962 93 + 6375548 GGCUUAAUGGCCAUCUUUUGCACUCAGAGUAAUGAUUUUGCCAUUGGUUAUCCCAUAGGUAAGU----AAGGCCAGAAGUAGCAAUCUUUAGGAAAU------------- .(((...((((((((..((((.......)))).)))((((((..(((.....)))..)))))).----..))))).....)))..............------------- ( -22.60, z-score = -0.62, R) >droVir3.scaffold_13049 18402402 87 - 25233164 GGCCUGAUGGCCAUUUUCUGCACGCAGAGCAACGACUUUGCCAUUGGCUAUCCCAUAGGUAAGCC---------GCAUCCUGGUUAUUGUUGUUCA-------------- ((((....))))..............(((((((((....((((.(((((..((....))..))))---------).....))))..))))))))).-------------- ( -29.00, z-score = -1.80, R) >droGri2.scaffold_15110 5976768 89 + 24565398 GGCGUCAUGGCCAUCUUUUGUACUCAGAGUAACGAUUUUGCCAUUGGUUAUCCCAUAGGUAAAC----AAUUGUA----UAGAAUUUCCCAAGCCAU------------- (((....(((.....((((((((((........)).((((((..(((.....)))..)))))).----....)))----)))))....))).)))..------------- ( -18.80, z-score = -0.96, R) >consensus GGCCUGAUGGCCAUAUUCUGCACCCAGAGCAAUGACUUUGCCAUCGGCUAUCCCAUAGGUGAGUU____UCAUAAGAACCUGAAUAUUGCAUCCAAA_____________ ((((....))))...(((((....))))).......((((((((.((....)).)).))))))............................................... (-12.02 = -10.92 + -1.10)

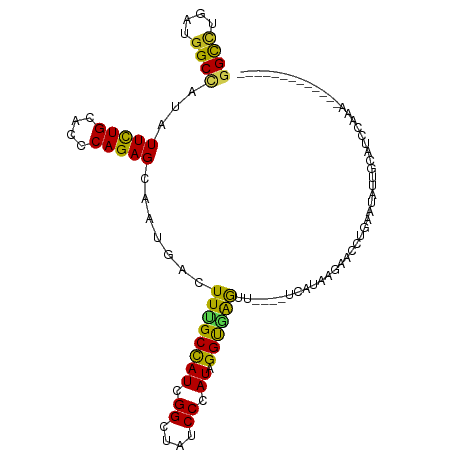

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:07 2011