
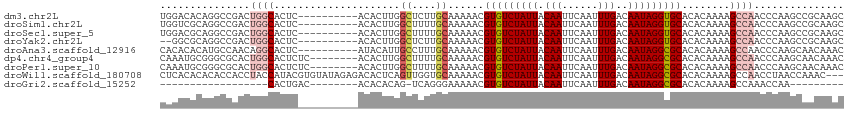
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,569,354 – 5,569,490 |
| Length | 136 |
| Max. P | 0.720213 |

| Location | 5,569,354 – 5,569,458 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.38 |
| Shannon entropy | 0.43328 |
| G+C content | 0.45608 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -12.42 |
| Energy contribution | -12.98 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.720213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5569354 104 + 23011544 UGGACACAGGCCGACUGGCACUC----------ACACUUGGCUCUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGUGCACACAAAAGCCAACCCAAGCCGCAAGC ........((((((.((......----------.)).))))))(((((......(..((((((.(((......)))..))))))..)........((........)).))))). ( -22.60, z-score = -0.07, R) >droSim1.chr2L 5364282 104 + 22036055 UGGUCGCAGGCCGACUGGCACUC----------ACACUUGGCUUUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGUGCACACAAAAGCCAACCCAAGCCGCAAGC .(((((.....)))))(((....----------....((((((((((.......(..((((((.(((......)))..))))))..)...)))))))))).....)))...... ( -26.22, z-score = -0.59, R) >droSec1.super_5 3647803 104 + 5866729 UGGACGCAGGCCGACUGGCACUC----------ACACUUGGCUUUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGUGCACACAAAAGCCAACCCAAGCCGCAAGC (((..(.(((((....))).)))----------....((((((((((.......(..((((((.(((......)))..))))))..)...)))))))))).)))...(....). ( -25.50, z-score = -0.58, R) >droYak2.chr2L 8699809 102 - 22324452 --GGCGCAGGCCGACUGGCACUC----------ACACUUGGCUCUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGUGCACACAAAAGCCAACCCAAGCCGCAAGC --((((((((((((.((......----------.)).))))))..)))......(..((((((.(((......)))..))))))..)........))).........(....). ( -26.20, z-score = -0.77, R) >droAna3.scaffold_12916 5157257 104 - 16180835 CACACACAUGCCAACAGGCACUC----------AUACAUUGCCUUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGCGCACACAAAAGCCAACCCAAGCAACAAAC ........(((.....(((....----------.....((((....))))....(((((((((.(((......)))..)))))))))........))).......)))...... ( -19.60, z-score = -2.19, R) >dp4.chr4_group4 3262648 106 + 6586962 CAAAUGCGGGCGCACUGGCACUCUC--------ACACUUGGCUUUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGCGCACACAAAAGCCAACCCAAGCAACAAAC ....((((((.((....)).)))..--------....((((((((((.......(((((((((.(((......)))..)))))))))...)))))))))).....)))...... ( -29.30, z-score = -3.30, R) >droPer1.super_10 2284017 106 + 3432795 CAAAUGCGGGCGCACUGGCACUCUC--------ACACUUGGCUUUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGCGCACACAAAAGCCAACCCAAGCAACAAAC ....((((((.((....)).)))..--------....((((((((((.......(((((((((.(((......)))..)))))))))...)))))))))).....)))...... ( -29.30, z-score = -3.30, R) >droWil1.scaffold_180708 8720773 111 + 12563649 CUCACACACACCACCUACCAUACGUGUAUAGAGACACUCAGUUGGUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGCGCACACAAAAGCCAACCUAACCAAAC--- (((..((((..............))))...))).......((((((........(((((((((.(((......)))..)))))))))........))))))..........--- ( -22.13, z-score = -2.21, R) >droGri2.scaffold_15252 14124348 78 - 17193109 ------------------CACUGAC--------ACACACAG-UCAGGGAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGCGCACACAAAAGCCAAACCAA--------- ------------------..(((((--------.......)-))))........(((((((((.(((......)))..)))))))))..................--------- ( -17.20, z-score = -3.20, R) >consensus CAGACGCAGGCCGACUGGCACUC__________ACACUUGGCUUUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGCGCACACAAAAGCCAACCCAAGCAACAAGC ...............((((.....................((....))......(((((((((.(((......)))..)))))))))........))))............... (-12.42 = -12.98 + 0.56)

| Location | 5,569,378 – 5,569,490 |
|---|---|
| Length | 112 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.88 |
| Shannon entropy | 0.38663 |
| G+C content | 0.44831 |
| Mean single sequence MFE | -19.87 |
| Consensus MFE | -11.72 |
| Energy contribution | -11.47 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.506611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5569378 112 + 23011544 CACUUGGCUCUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGUGCACACAAAAGCCAACCCAAGCCGCAAGCCAACCAUCCAGCCAUCCAACC----GCACUAAGCCA ....((((((((((......(..((((((.(((......)))..))))))..)........((........)).)))))..........))))).......----((.....)).. ( -20.00, z-score = -0.94, R) >droSim1.chr2L 5364306 112 + 22036055 CACUUGGCUUUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGUGCACACAAAAGCCAACCCAAGCCGCAAGCCAACCAUCCAGCCAUCCAACC----GCACUAAGCCA ....((((((.(((......(..((((((.(((......)))..))))))..)..................(.(....).)....................----)))..)))))) ( -20.80, z-score = -1.12, R) >droSec1.super_5 3647827 112 + 5866729 CACUUGGCUUUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGUGCACACAAAAGCCAACCCAAGCCGCAAGCCAACCAUCCAGCCAUCCAACC----GCACUAAGCCA ....((((((.(((......(..((((((.(((......)))..))))))..)..................(.(....).)....................----)))..)))))) ( -20.80, z-score = -1.12, R) >droYak2.chr2L 8699831 116 - 22324452 CACUUGGCUCUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGUGCACACAAAAGCCAACCCAAGCCGCAAGCCAACCAUCCAACCAUCCAACCAACCGCACUAAGCCA ...((((((..(((......(..((((((.(((......)))..))))))..)........((........)).)))))))))......................((.....)).. ( -19.30, z-score = -1.25, R) >droAna3.scaffold_12916 5157281 104 - 16180835 UACAUUGCCUUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGCGCACACAAAAGCCAACCCAAGCAACAAACCAACCAUUCAACC------------GCGCUAAGCUG ....((((....))))....(((((((((.(((......)))..)))))))))........((........))....................------------((.....)).. ( -15.40, z-score = -0.64, R) >dp4.chr4_group4 3262674 116 + 6586962 CACUUGGCUUUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGCGCACACAAAAGCCAACCCAAGCAACAAACCAAACAUCCAACCAUCCAUUCAACCGCACUAAGCUA ...((((((((((.......(((((((((.(((......)))..)))))))))...)))))))))).......................................((.....)).. ( -22.00, z-score = -3.38, R) >droPer1.super_10 2284043 116 + 3432795 CACUUGGCUUUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGCGCACACAAAAGCCAACCCAAGCAACAAACCAAACAUCCAACCAUCCAUUCAACCGCACUAAGCUA ...((((((((((.......(((((((((.(((......)))..)))))))))...)))))))))).......................................((.....)).. ( -22.00, z-score = -3.38, R) >droWil1.scaffold_180708 8720807 98 + 12563649 CACUCAGUUGGUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGCGCACACAAAAGCCAACCUAAC---CAAACCAAA---CCAACC------------GCACUGAGUCU .(((((((((((........(((((((((.(((......)))..))))))))).......((.....))...---.........---...)))------------).))))))).. ( -22.00, z-score = -2.78, R) >droMoj3.scaffold_6500 18448431 95 - 32352404 CAGUCGCCU--UGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGCGCACACAAAAGCCAAGCCAAG---CCAGCCAG----CCAGCC------------GCACUGUGUUG ((((((.((--.((......(((((((((.(((......)))..)))))))))........((........)---)......)----).)).)------------).))))..... ( -20.10, z-score = 0.05, R) >droVir3.scaffold_12963 16686418 86 - 20206255 CAUUCGCAU--UGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGCGCACACAAAAGCCAAAGCAA----------------CCAGCC------------GCACUGAGCUG ......((.--(((......(((((((((.(((......)))..)))))))))........((....))..----------------......------------))).))..... ( -16.30, z-score = -0.38, R) >consensus CACUUGGCUUUUGCAAAAACGUGUCUAUUACAAUUCAAUUUGACAAUAGGCGCACACAAAAGCCAACCCAAGCCGCAAGCCAACCAUCCAACCAUCCA__C____GCACUAAGCCA ....(((((..(((......(((((((((.(((......)))..)))))))))........(......)....................................)))...))))) (-11.72 = -11.47 + -0.25)

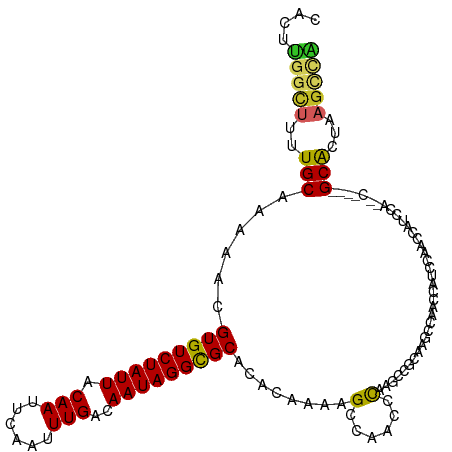

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:22 2011