| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,576,076 – 17,576,168 |
| Length | 92 |
| Max. P | 0.986669 |

| Location | 17,576,076 – 17,576,168 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 79.10 |
| Shannon entropy | 0.34366 |
| G+C content | 0.53484 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -18.49 |
| Energy contribution | -19.13 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
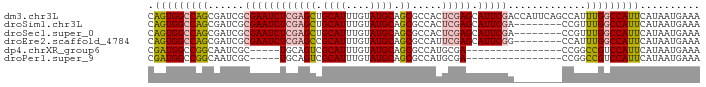
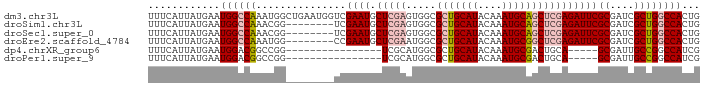
>dm3.chr3L 17576076 92 + 24543557 CAGUGGCCAGCGAUCGCGAAUCUCGAGCUGCAUUUGUAUGCAGCGCCACUCGAGCAUUCGACCAUUCAGCCAUUUGGCCAUUCAUAAUGAAA .((((((((((((...(((((((((((((((((....))))))).....))))).))))).....)).))....)))))))).......... ( -33.80, z-score = -3.10, R) >droSim1.chr3L 16914202 84 + 22553184 CAGUGGCCAGCGAUCGCGAAUCUCGAGCUGCAUUUGUAUGCAGCGCCACUCGAGCAUUCGA--------CCGUUUGGCCAUUCAUAAUGAAA .(((((((((((....(((((((((((((((((....))))))).....))))).))))).--------.)).))))))))).......... ( -35.50, z-score = -3.98, R) >droSec1.super_0 9622821 84 + 21120651 CAGUGGCCAGCGAUCGCGAAUCUCGAGCUGCAUUUGUAUGCAGCGCCACUCGAGCAUUCGA--------CCGUUUGGCCAUUCAUAAUGAAA .(((((((((((....(((((((((((((((((....))))))).....))))).))))).--------.)).))))))))).......... ( -35.50, z-score = -3.98, R) >droEre2.scaffold_4784 9483890 84 - 25762168 CAGUGGCCAGCGAUCGCGAAUCUCGAGCCGCAUUUGUAUGCAGCGCCAUUCGAGCAUUCGG--------CCAUUUGGCCAUUCAUAAUGAAA .((((((((((....))(((((((((((.((((....)))).)).....))))).))))..--------.....)))))))).......... ( -28.30, z-score = -1.36, R) >dp4.chrXR_group6 2242661 71 + 13314419 CGAUGGCCGGCAAUCGC-----UGCAGUCGCAUUUGUAUGCAGCGCCAUGCGA----------------CCGGCCGUCCAUUCAUAAUGAAA .(((((((((...((((-----.((.((.((((....)))).))))...))))----------------)))))))))..(((.....))). ( -29.80, z-score = -2.59, R) >droPer1.super_9 533187 71 + 3637205 CGAUGGCCGGCAAUCGC-----UGCAGUCGCAUUUGUAUGCAGCGCCAUGCGA----------------CCGGCCGUCCAUUCAUAAUGAAA .(((((((((...((((-----.((.((.((((....)))).))))...))))----------------)))))))))..(((.....))). ( -29.80, z-score = -2.59, R) >consensus CAGUGGCCAGCGAUCGCGAAUCUCGAGCCGCAUUUGUAUGCAGCGCCACUCGAGCAUUCGA________CCGUUUGGCCAUUCAUAAUGAAA .((((((((((....))(((((((((((.((((....)))).)).....))))).))))...............)))))))).......... (-18.49 = -19.13 + 0.64)
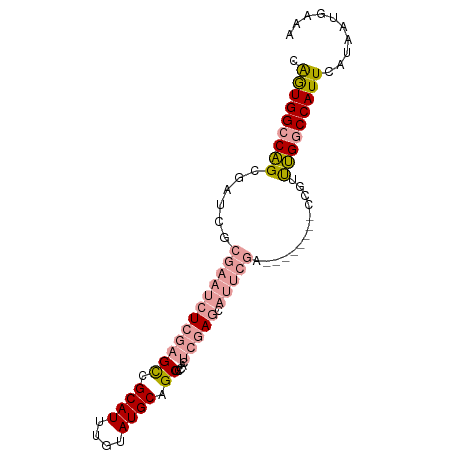
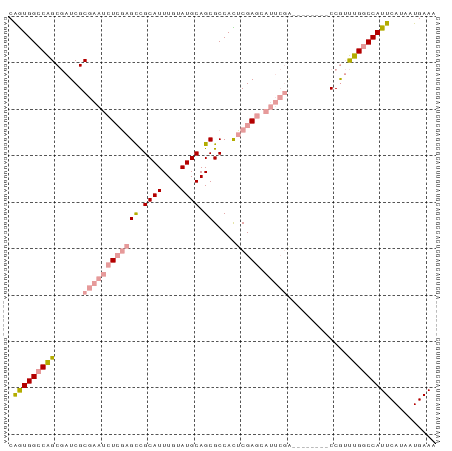
| Location | 17,576,076 – 17,576,168 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 79.10 |
| Shannon entropy | 0.34366 |
| G+C content | 0.53484 |
| Mean single sequence MFE | -30.95 |
| Consensus MFE | -18.49 |
| Energy contribution | -19.88 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
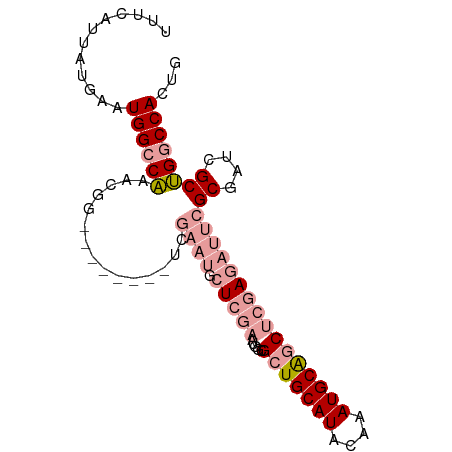
>dm3.chr3L 17576076 92 - 24543557 UUUCAUUAUGAAUGGCCAAAUGGCUGAAUGGUCGAAUGCUCGAGUGGCGCUGCAUACAAAUGCAGCUCGAGAUUCGCGAUCGCUGGCCACUG ............((((((...((((....))))((((.(((((.....(((((((....))))))))))))))))((....))))))))... ( -33.00, z-score = -1.94, R) >droSim1.chr3L 16914202 84 - 22553184 UUUCAUUAUGAAUGGCCAAACGG--------UCGAAUGCUCGAGUGGCGCUGCAUACAAAUGCAGCUCGAGAUUCGCGAUCGCUGGCCACUG ............((((((..(((--------(((....(((((.....(((((((....)))))))))))).....)))))).))))))... ( -34.70, z-score = -3.55, R) >droSec1.super_0 9622821 84 - 21120651 UUUCAUUAUGAAUGGCCAAACGG--------UCGAAUGCUCGAGUGGCGCUGCAUACAAAUGCAGCUCGAGAUUCGCGAUCGCUGGCCACUG ............((((((..(((--------(((....(((((.....(((((((....)))))))))))).....)))))).))))))... ( -34.70, z-score = -3.55, R) >droEre2.scaffold_4784 9483890 84 + 25762168 UUUCAUUAUGAAUGGCCAAAUGG--------CCGAAUGCUCGAAUGGCGCUGCAUACAAAUGCGGCUCGAGAUUCGCGAUCGCUGGCCACUG ............((((((.((.(--------(.((((.(((((.....(((((((....)))))))))))))))))).))...))))))... ( -30.70, z-score = -2.20, R) >dp4.chrXR_group6 2242661 71 - 13314419 UUUCAUUAUGAAUGGACGGCCGG----------------UCGCAUGGCGCUGCAUACAAAUGCGACUGCA-----GCGAUUGCCGGCCAUCG ..............((.((((((----------------(.......(((((((..(......)..))))-----)))...))))))).)). ( -26.30, z-score = -1.50, R) >droPer1.super_9 533187 71 - 3637205 UUUCAUUAUGAAUGGACGGCCGG----------------UCGCAUGGCGCUGCAUACAAAUGCGACUGCA-----GCGAUUGCCGGCCAUCG ..............((.((((((----------------(.......(((((((..(......)..))))-----)))...))))))).)). ( -26.30, z-score = -1.50, R) >consensus UUUCAUUAUGAAUGGCCAAACGG________UCGAAUGCUCGAAUGGCGCUGCAUACAAAUGCAGCUCGAGAUUCGCGAUCGCUGGCCACUG ............((((((...............((((.(((((.....(((((((....))))))))))))))))((....))))))))... (-18.49 = -19.88 + 1.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:04 2011