
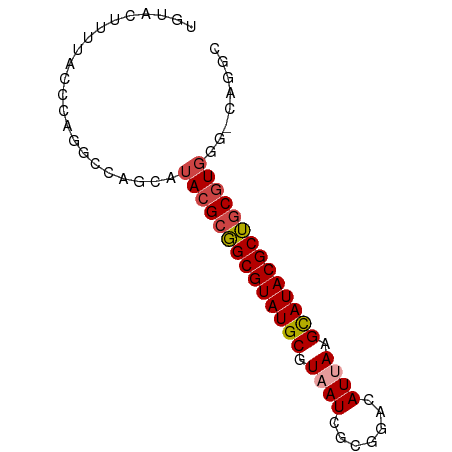
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,531,508 – 17,531,583 |
| Length | 75 |
| Max. P | 0.998632 |

| Location | 17,531,508 – 17,531,583 |
|---|---|
| Length | 75 |
| Sequences | 6 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 73.56 |
| Shannon entropy | 0.50931 |
| G+C content | 0.54508 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -21.85 |
| Energy contribution | -22.27 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.998632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
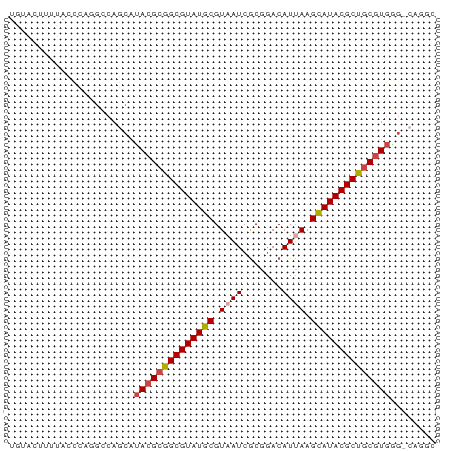
>dm3.chr3L 17531508 75 + 24543557 UAUACUUUUACCCAGGCCAGCGUACGCGGCGUAUGCGUAAUCGCGGACAUUAAGCAUACGCUGCGUGGGUCAGGC ...............(((.((.((((((((((((((.((((.......)))).)))))))))))))).))..))) ( -32.10, z-score = -2.85, R) >droEre2.scaffold_4784 9438772 67 - 25762168 UGAAAUUUUACCCAGGCCAGCAUACGCGGCGUAUGCGUAAUCGCGGACAUCAAGCAUACGCUGCGUG-------- ......................((((((((((((((.................))))))))))))))-------- ( -23.33, z-score = -1.79, R) >droYak2.chr3L 7656380 61 - 24197627 --------------UGCCAGCAUACGCGGCGUAUGCGUAAUCGCGCACAUCAAGCAUACGCUGCGUGAGCCAGGC --------------.(((.((.((((((((((((((.................)))))))))))))).))..))) ( -30.43, z-score = -3.32, R) >droSim1.chr3L 16863484 75 + 22553184 UGUACUUUUACCCAGGCCAGCAUACGCGGCGUAUGCGUAAUCGCGGACAUUAAGCAUACGCUGCGUGGGUCAGGC ...........((.((((....((((((((((((((.((((.......)))).)))))))))))))))))).)). ( -30.90, z-score = -2.50, R) >droSec1.super_0 9576048 69 + 21120651 UGUACUUUUACCCUGCCCAGCAUACGCGGCGUAUGCGUAAUCGCGGACAUUAAGCAUACGCUGCGUGGG------ ..........((((((...))).(((((((((((((.((((.......)))).))))))))))))))))------ ( -27.90, z-score = -2.42, R) >droMoj3.scaffold_6680 19529265 68 - 24764193 -------UAGAUGUUGGCAGCGAACGUUGCGUAUGCGUAAUCACAGACAUUAAGUAUACGCAUCCUAUGGCCAAC -------.....((((((.........(((((((((.((((.......)))).))))))))).......)))))) ( -21.39, z-score = -2.38, R) >consensus UGUACUUUUACCCAGGCCAGCAUACGCGGCGUAUGCGUAAUCGCGGACAUUAAGCAUACGCUGCGUGGG_CAGGC ......................((((((((((((((.((((.......)))).))))))))))))))........ (-21.85 = -22.27 + 0.42)

| Location | 17,531,508 – 17,531,583 |
|---|---|
| Length | 75 |
| Sequences | 6 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 73.56 |
| Shannon entropy | 0.50931 |
| G+C content | 0.54508 |
| Mean single sequence MFE | -25.85 |
| Consensus MFE | -18.27 |
| Energy contribution | -18.55 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.986083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
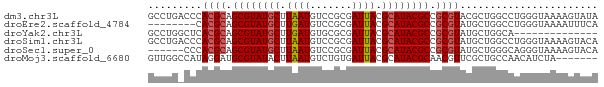
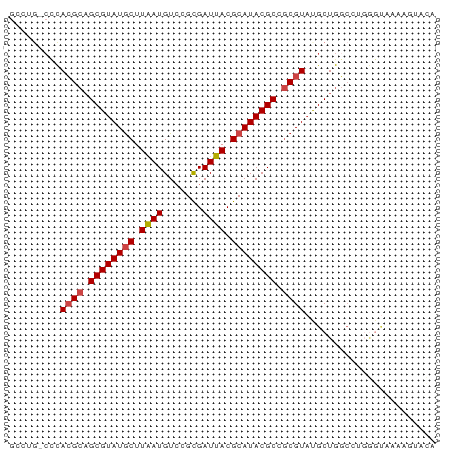
>dm3.chr3L 17531508 75 - 24543557 GCCUGACCCACGCAGCGUAUGCUUAAUGUCCGCGAUUACGCAUACGCCGCGUACGCUGGCCUGGGUAAAAGUAUA ..((.(((((.(((((((((((....(((..(((....)))..)))..))))))))).)).)))))...)).... ( -29.30, z-score = -2.17, R) >droEre2.scaffold_4784 9438772 67 + 25762168 --------CACGCAGCGUAUGCUUGAUGUCCGCGAUUACGCAUACGCCGCGUAUGCUGGCCUGGGUAAAAUUUCA --------.((((.((((((((.(((((....).)))).)))))))).)))).((((......))))........ ( -22.30, z-score = -0.77, R) >droYak2.chr3L 7656380 61 + 24197627 GCCUGGCUCACGCAGCGUAUGCUUGAUGUGCGCGAUUACGCAUACGCCGCGUAUGCUGGCA-------------- (((.(((..((((.((((((((.(((((....).)))).)))))))).))))..)))))).-------------- ( -31.00, z-score = -2.85, R) >droSim1.chr3L 16863484 75 - 22553184 GCCUGACCCACGCAGCGUAUGCUUAAUGUCCGCGAUUACGCAUACGCCGCGUAUGCUGGCCUGGGUAAAAGUACA ..((.(((((.(((((((((((....(((..(((....)))..)))..))))))))).)).)))))...)).... ( -27.50, z-score = -1.60, R) >droSec1.super_0 9576048 69 - 21120651 ------CCCACGCAGCGUAUGCUUAAUGUCCGCGAUUACGCAUACGCCGCGUAUGCUGGGCAGGGUAAAAGUACA ------(((((((.((((((((.(((((....).)))).)))))))).)))).(((...)))))).......... ( -26.00, z-score = -1.46, R) >droMoj3.scaffold_6680 19529265 68 + 24764193 GUUGGCCAUAGGAUGCGUAUACUUAAUGUCUGUGAUUACGCAUACGCAACGUUCGCUGCCAACAUCUA------- ((((((....((((((((((.(.((((.......)))).).)))))))...)))...)))))).....------- ( -19.00, z-score = -1.33, R) >consensus GCCUG_CCCACGCAGCGUAUGCUUAAUGUCCGCGAUUACGCAUACGCCGCGUAUGCUGGCCUGGGUAAAAGUACA .........((((.((((((((.((((.......)))).)))))))).))))....................... (-18.27 = -18.55 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:02 2011