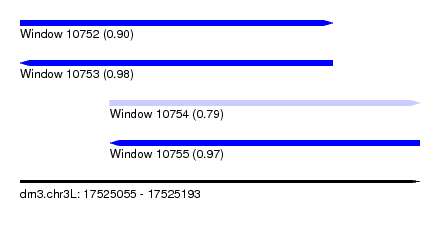
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,525,055 – 17,525,193 |
| Length | 138 |
| Max. P | 0.979714 |

| Location | 17,525,055 – 17,525,163 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Shannon entropy | 0.27165 |
| G+C content | 0.37318 |
| Mean single sequence MFE | -26.66 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.902150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17525055 108 + 24543557 UAAAGAAAAAGUAGCUUUUUUGCGGAUUUUUUAAAAUCGAAUAACAUUUCUUGACUUGCACUUUCCGAAGAGCAGAAAAGUGCCAAGAAAAGUAUUAGGAAGCCGCCA ..........(..((((((((((.(((((....)))))........(((((((....(((((((.(........).)))))))))))))).)))..)))))))..).. ( -23.80, z-score = -1.32, R) >droYak2.chr3L 7649750 103 - 24197627 UAAAGAAGAAAUAGCCUUCUUGCGGAUUCUAUAAAAGCUAAACGCAACUCUUGGUUUGCACUUUUC-----GCAGAAGGGUGCCAAGAAAAGUAUUAUGAAGCCGCCA ..((((((.......))))))((((.(((.((((..((.....))...(((((....(((((((((-----...))))))))))))))......))))))).)))).. ( -28.10, z-score = -1.69, R) >droSim1.chr3L 16857099 108 + 22553184 UAAAGAAAAAGUAGCCUUUUUGCGGAUUUUUUAAAAUCCAAACACAUUUCUUGACUUGCACUUUCCGAAGAGCAGAAAAGUGCCAAGAAAAGUAUUUGGUAGCCGCCA .....((((((....))))))((((............(((((.((.(((((((....(((((((.(........).)))))))))))))).)).)))))...)))).. ( -26.86, z-score = -2.40, R) >droSec1.super_0 9569783 102 + 21120651 ------UAAAGUAGCCUUUUUGCGGAUUUUUUAAAAUCUAAACACAUUUCUUGACUUGCACUUUUCAAAGAGCCGAAAAGUGCCAAGAAAAGUAUUAGGUAGCCGCCA ------....((.((((...(((((((((....)))))).......(((((((....(((((((((........)))))))))))))))).)))..)))).))..... ( -27.90, z-score = -3.21, R) >consensus UAAAGAAAAAGUAGCCUUUUUGCGGAUUUUUUAAAAUCUAAACACAUUUCUUGACUUGCACUUUCCGAAGAGCAGAAAAGUGCCAAGAAAAGUAUUAGGAAGCCGCCA .....((((((....))))))((((....(((((.........((.(((((((....(((((((((........)))))))))))))))).)).)))))...)))).. (-19.26 = -19.82 + 0.56)

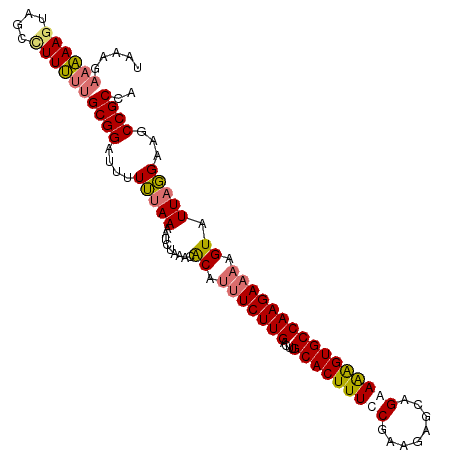
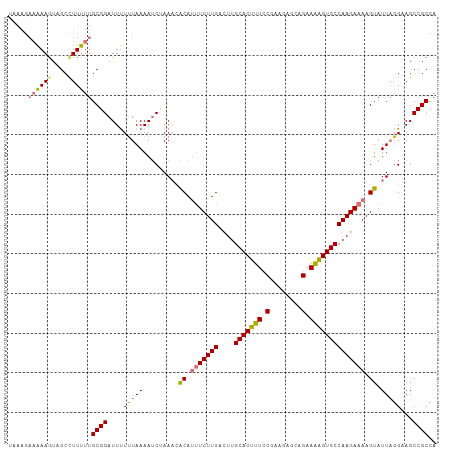
| Location | 17,525,055 – 17,525,163 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Shannon entropy | 0.27165 |
| G+C content | 0.37318 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -21.33 |
| Energy contribution | -22.70 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17525055 108 - 24543557 UGGCGGCUUCCUAAUACUUUUCUUGGCACUUUUCUGCUCUUCGGAAAGUGCAAGUCAAGAAAUGUUAUUCGAUUUUAAAAAAUCCGCAAAAAAGCUACUUUUUCUUUA .((..((((..(((((..((((((((((((((((........)))))))))....))))))))))))...(((((....))))).......))))..))......... ( -23.20, z-score = -1.70, R) >droYak2.chr3L 7649750 103 + 24197627 UGGCGGCUUCAUAAUACUUUUCUUGGCACCCUUCUGC-----GAAAAGUGCAAACCAAGAGUUGCGUUUAGCUUUUAUAGAAUCCGCAAGAAGGCUAUUUCUUCUUUA ..((((.(((....((((((((...(((......)))-----))))))))......((((((((....))))))))...))).))))(((((((.....))))))).. ( -26.60, z-score = -1.28, R) >droSim1.chr3L 16857099 108 - 22553184 UGGCGGCUACCAAAUACUUUUCUUGGCACUUUUCUGCUCUUCGGAAAGUGCAAGUCAAGAAAUGUGUUUGGAUUUUAAAAAAUCCGCAAAAAGGCUACUUUUUCUUUA ..((((...((((((((.((((((((((((((((........)))))))))....))))))).))))))))((((....))))))))((((((....))))))..... ( -34.90, z-score = -4.55, R) >droSec1.super_0 9569783 102 - 21120651 UGGCGGCUACCUAAUACUUUUCUUGGCACUUUUCGGCUCUUUGAAAAGUGCAAGUCAAGAAAUGUGUUUAGAUUUUAAAAAAUCCGCAAAAAGGCUACUUUA------ ..((((....(((((((.((((((((((((((((((....)))))))))))....))))))).))).))))((((....))))))))..((((....)))).------ ( -28.00, z-score = -2.68, R) >consensus UGGCGGCUACCUAAUACUUUUCUUGGCACUUUUCUGCUCUUCGAAAAGUGCAAGUCAAGAAAUGUGUUUAGAUUUUAAAAAAUCCGCAAAAAGGCUACUUUUUCUUUA ..(((.......(((((.((((((((((((((((........)))))))))....))))))).)))))..(((((....))))))))((((((....))))))..... (-21.33 = -22.70 + 1.37)

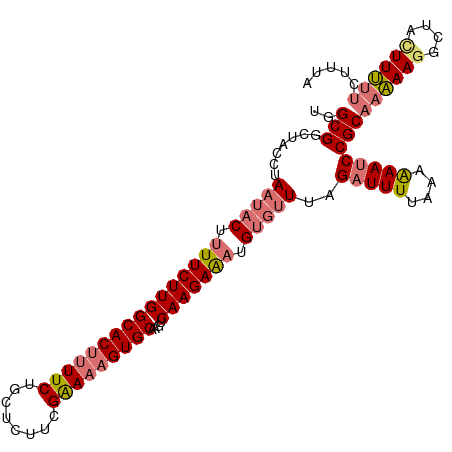
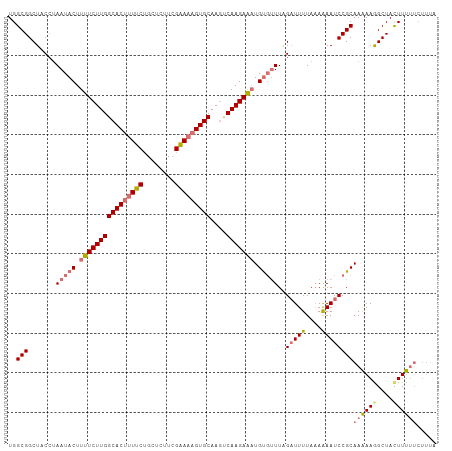
| Location | 17,525,086 – 17,525,193 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.32 |
| Shannon entropy | 0.52614 |
| G+C content | 0.38951 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -9.56 |
| Energy contribution | -9.24 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.793824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17525086 107 + 24543557 UAAAAUCGAAUAACAUUUCUUGACUUGCACUUUCCGAAGAGCAGAAAAGUGCCAAGAAAAGUAUUAGGAAGCCGCCA-GAAAAUAAACUGGCAUCAUCGUUGUUUACU--- .......(((((((.(((((((....(((((((.(........).))))))))))))))..............((((-(........)))))......)))))))...--- ( -27.40, z-score = -3.17, R) >droYak2.chr3L 7649781 102 - 24197627 UAAAAGCUAAACGCAACUCUUGGUUUGCACUUUUC-----GCAGAAGGGUGCCAAGAAAAGUAUUAUGAAGCCGCCA-GAAAAUAAACUGGCAUCAUUGUUGUUUACU--- ....((.(((((((((.(((((....(((((((((-----...))))))))))))))................((((-(........)))))....))).))))))))--- ( -25.60, z-score = -1.32, R) >droSim1.chr3L 16857130 107 + 22553184 UAAAAUCCAAACACAUUUCUUGACUUGCACUUUCCGAAGAGCAGAAAAGUGCCAAGAAAAGUAUUUGGUAGCCGCCA-GAAAAUAAACUGGCAUCAUCGUUGUUUGCU--- ......(((((.((.(((((((....(((((((.(........).)))))))))))))).)).))))).....((((-(........)))))................--- ( -28.50, z-score = -2.87, R) >droSec1.super_0 9569808 107 + 21120651 UAAAAUCUAAACACAUUUCUUGACUUGCACUUUUCAAAGAGCCGAAAAGUGCCAAGAAAAGUAUUAGGUAGCCGCCA-GAAAAUAAACUGGCAUCAUUGUUGUUUGCU--- ....((((((..((.(((((((....(((((((((........)))))))))))))))).)).))))))....((((-(........)))))................--- ( -30.00, z-score = -3.60, R) >droWil1.scaffold_181009 207336 106 - 3585778 CAAAGUUUAGUCA-ACCUCUUCGUUAAAGGAC----AGGCCCAAAAGAGUCCCCAAAAAAGUAUGCAACAUGCGUGAUGGUAAAAGAUUCGUGAGGUUUUCGUGACCAUGA .........((((-(((((.........((..----....))....(((((..........(((((.....))))).........)))))..))))).....))))..... ( -19.41, z-score = 0.36, R) >consensus UAAAAUCUAAACACAUUUCUUGACUUGCACUUUCC_AAGAGCAGAAAAGUGCCAAGAAAAGUAUUAGGAAGCCGCCA_GAAAAUAAACUGGCAUCAUUGUUGUUUACU___ ..................(((..((((((((((.(........).)))))).))))..)))............((((...........))))................... ( -9.56 = -9.24 + -0.32)

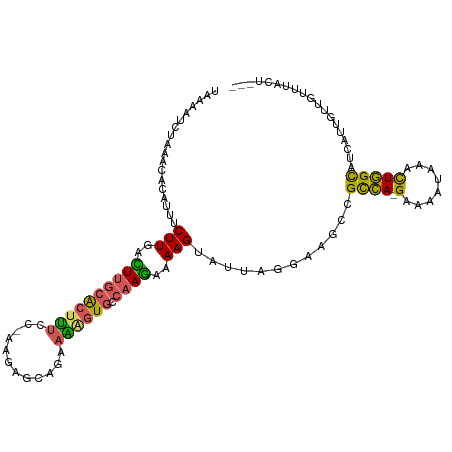
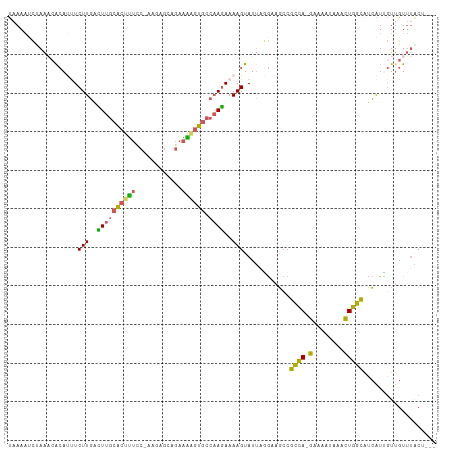
| Location | 17,525,086 – 17,525,193 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 70.32 |
| Shannon entropy | 0.52614 |
| G+C content | 0.38951 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -13.00 |
| Energy contribution | -14.92 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17525086 107 - 24543557 ---AGUAAACAACGAUGAUGCCAGUUUAUUUUC-UGGCGGCUUCCUAAUACUUUUCUUGGCACUUUUCUGCUCUUCGGAAAGUGCAAGUCAAGAAAUGUUAUUCGAUUUUA ---....((((..((...((((((........)-)))))...))........((((((((((((((((........)))))))))....)))))))))))........... ( -28.00, z-score = -2.50, R) >droYak2.chr3L 7649781 102 + 24197627 ---AGUAAACAACAAUGAUGCCAGUUUAUUUUC-UGGCGGCUUCAUAAUACUUUUCUUGGCACCCUUCUGC-----GAAAAGUGCAAACCAAGAGUUGCGUUUAGCUUUUA ---..(((((....((((((((((........)-)))))...))))...(((((((...(((......)))-----)))))))((((........)))))))))....... ( -22.60, z-score = -0.54, R) >droSim1.chr3L 16857130 107 - 22553184 ---AGCAAACAACGAUGAUGCCAGUUUAUUUUC-UGGCGGCUACCAAAUACUUUUCUUGGCACUUUUCUGCUCUUCGGAAAGUGCAAGUCAAGAAAUGUGUUUGGAUUUUA ---(((...((....)).((((((........)-)))))))).((((((((.((((((((((((((((........)))))))))....))))))).))))))))...... ( -38.20, z-score = -4.91, R) >droSec1.super_0 9569808 107 - 21120651 ---AGCAAACAACAAUGAUGCCAGUUUAUUUUC-UGGCGGCUACCUAAUACUUUUCUUGGCACUUUUCGGCUCUUUGAAAAGUGCAAGUCAAGAAAUGUGUUUAGAUUUUA ---(((...((....)).((((((........)-))))))))..(((((((.((((((((((((((((((....)))))))))))....))))))).))).))))...... ( -32.50, z-score = -3.60, R) >droWil1.scaffold_181009 207336 106 + 3585778 UCAUGGUCACGAAAACCUCACGAAUCUUUUACCAUCACGCAUGUUGCAUACUUUUUUGGGGACUCUUUUGGGCCU----GUCCUUUAACGAAGAGGU-UGACUAAACUUUG ...((((((.....(((((...................((.....))...........(((.(((....))))))----.............)))))-))))))....... ( -19.90, z-score = 0.22, R) >consensus ___AGCAAACAACAAUGAUGCCAGUUUAUUUUC_UGGCGGCUACCUAAUACUUUUCUUGGCACUUUUCUGCUCUU_GAAAAGUGCAAGUCAAGAAAUGUGUUUAGAUUUUA ..................(((((...........))))).......(((((.((((((((((((((((........)))))))))....))))))).)))))......... (-13.00 = -14.92 + 1.92)

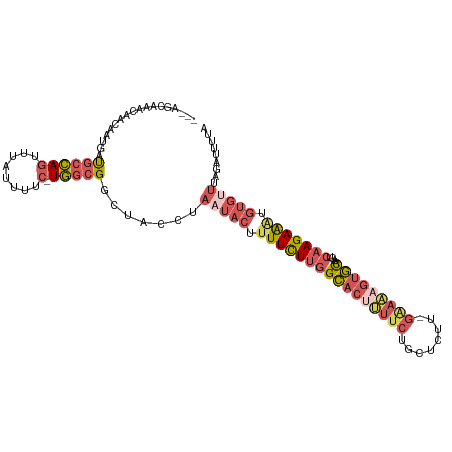
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:34:00 2011