| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,517,291 – 17,517,392 |
| Length | 101 |
| Max. P | 0.999503 |

| Location | 17,517,291 – 17,517,392 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 51.15 |
| Shannon entropy | 0.88435 |
| G+C content | 0.48980 |
| Mean single sequence MFE | -28.61 |
| Consensus MFE | -15.80 |
| Energy contribution | -16.32 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
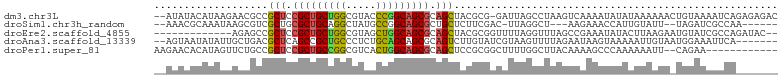
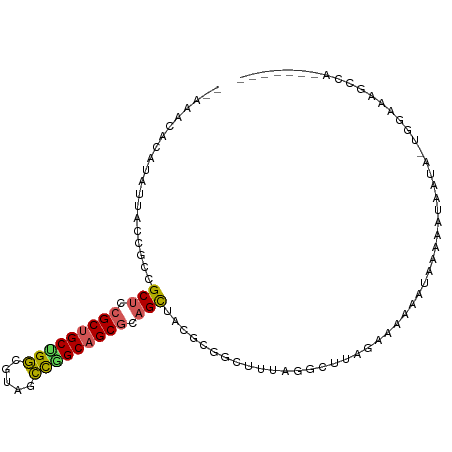
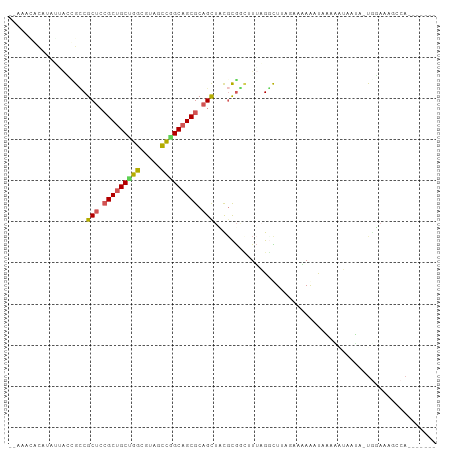
>dm3.chr3L 17517291 101 + 24543557 GUCUCUCUGAUUUUACAGUUUUUUAUAUAUUUUGACUUAGGCUAAUC-CGCGUAGCUGCGCUGCCGGGUACGCCAGCAGCGGAGCGGCGUUCUUAUGUAUAU-- ......(((......))).....(((((((...((....((.....)-)((((.(((.((((((.((.....)).)))))).))).))))))..))))))).-- ( -29.40, z-score = -0.79, R) >droSim1.chr3h_random 107918 90 - 1452968 ------UUGGCGAUCUA--AAUACAAUGGUUUCUU---AGCCUAA-GUCGAAGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUAUUUGCGUUU-- ------(((((......--........((((....---))))...-)))))...((.(((((((.(((...))).))))))).))(((((......))))).-- ( -31.73, z-score = -1.05, R) >droEre2.scaffold_4855 7528 89 + 242605 --GUAUCUGGCGAUACAUUCUUAAGUAUAUUUCGGCUAAACCUAAAACCGCGUAGCUGCGCUGCCAGCUACGCCAGCAGCGGAGCGGCUCU------------- --(((.((((((((((........))))....((((((..(........)..))))))...)))))).)))(((.((......)))))...------------- ( -25.10, z-score = 0.47, R) >droAna3.scaffold_13339 1856681 95 + 4599533 -------UGAAUUUCCAUUACAAUUUUUACUUAUUCUAAAACUUACGAUACAAGACUGCGCUGCUGCAGAGGGCAGCGGCUGAGCGUCAGCAAUAUAUUACU-- -------..............................................(((.((((((((((.....))))))))...)))))..............-- ( -19.00, z-score = -0.28, R) >droPer1.super_81 267482 90 - 277874 ------------UUCUG--AAUUUUUUGGGCUUUUGUAAGCCAAAAGCCGCGGAGCUGCGCUGCCAGUGACGCCGGCAGCGGAGCGGCAGAACUAUGUGUUCUU ------------.....--.........((((((((.....))))))))((...(((.(((((((.(.....).))))))).))).))(((((.....))))). ( -37.80, z-score = -2.01, R) >consensus _______UGACGUUCCA_UAAUAUAUUUAUUUCUUCUAAGCCUAAAGCCGCGGAGCUGCGCUGCCGGCGACGCCAGCAGCGGAGCGGCACUAAUAUGUGUUU__ ......................................................(((.((((((.((.....)).)))))).)))................... (-15.80 = -16.32 + 0.52)
| Location | 17,517,291 – 17,517,392 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 51.15 |
| Shannon entropy | 0.88435 |
| G+C content | 0.48980 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -18.66 |
| Energy contribution | -18.50 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
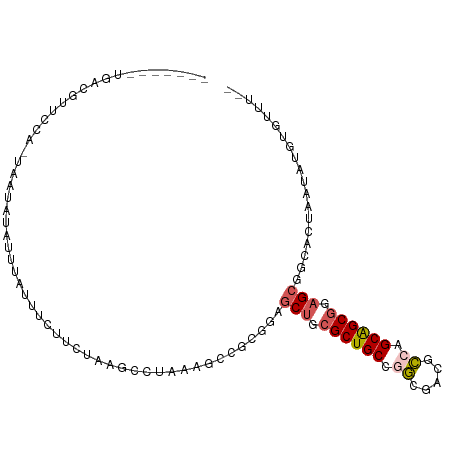
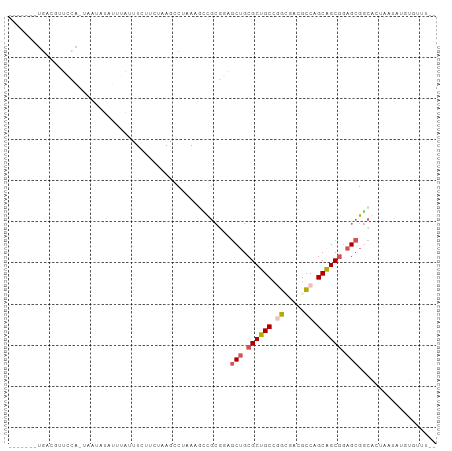
>dm3.chr3L 17517291 101 - 24543557 --AUAUACAUAAGAACGCCGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCG-GAUUAGCCUAAGUCAAAAUAUAUAAAAAACUGUAAAAUCAGAGAGAC --(((((........(((.(((.(((((((((.....))))))))).)))...)))-((((......))))....)))))......(((......)))...... ( -28.40, z-score = -2.11, R) >droSim1.chr3h_random 107918 90 + 1452968 --AAACGCAAAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCUUCGAC-UUAGGCU---AAGAAACCAUUGUAUU--UAGAUCGCCAA------ --..((((......)))).((.(((((((.(((...))).))))))).))........-...(((.---................--......)))..------ ( -28.50, z-score = -0.47, R) >droEre2.scaffold_4855 7528 89 - 242605 -------------AGAGCCGCUCCGCUGCUGGCGUAGCUGGCAGCGCAGCUACGCGGUUUUAGGUUUAGCCGAAAUAUACUUAAGAAUGUAUCGCCAGAUAC-- -------------...((.(((.((((((..((...))..)))))).)))...))..((((((((.((.......)).))))))))..(((((....)))))-- ( -30.40, z-score = -0.69, R) >droAna3.scaffold_13339 1856681 95 - 4599533 --AGUAAUAUAUUGCUGACGCUCAGCCGCUGCCCUCUGCAGCAGCGCAGUCUUGUAUCGUAAGUUUUAGAAUAAGUAAAAAUUGUAAUGGAAAUUCA------- --.(..(((((..((((.((((.....(((((.....)))))))))))))..)))))..)...(((((..((((.......))))..))))).....------- ( -21.10, z-score = -0.34, R) >droPer1.super_81 267482 90 + 277874 AAGAACACAUAGUUCUGCCGCUCCGCUGCCGGCGUCACUGGCAGCGCAGCUCCGCGGCUUUUGGCUUACAAAAGCCCAAAAAAUU--CAGAA------------ .(((((.....)))))((.(((.(((((((((.....))))))))).)))...))((((((((.....)))))))).........--.....------------ ( -38.00, z-score = -3.97, R) >consensus __AAACACAUAUUACCGCCGCUCCGCUGCUGGCGUAGCCGGCAGCGCAGCUACGCGGCUUUAGGCUUAGAAAAAAUAAAAAUAAUA_UGGAAAGCCA_______ ...................(((.(((((((((.....))))))))).)))...................................................... (-18.66 = -18.50 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:55 2011