| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,510,274 – 17,510,364 |
| Length | 90 |
| Max. P | 0.873975 |

| Location | 17,510,274 – 17,510,364 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 54.61 |
| Shannon entropy | 0.88734 |
| G+C content | 0.51549 |
| Mean single sequence MFE | -26.21 |
| Consensus MFE | -12.63 |
| Energy contribution | -12.81 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.42 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.873975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17510274 90 - 24543557 --AAGAACGCCGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGAUUAGCCUAAGUCA-AAAU-AUAUAAAAAACUGUAAAAUCAGAGA --.....(((.(((.(((((((((.....))))))))).)))...)))((((......)))).-....-..........(((......)))... ( -27.70, z-score = -1.85, R) >droAna3.scaffold_13339 1849457 84 - 4599533 --AUUGCUGACGCUCAGCCGCUGCCCUCUGCAGCAGCGCAGUCUUGUAUCGUAAGUUUUAGAAUAAGUAAAAAUUGUAAUGGAAAU-------- --...((((.((((.....(((((.....))))))))))))).((.(((..(((.(((((.......))))).)))..))).))..-------- ( -18.10, z-score = 0.22, R) >droYak2.chrX_random 539142 88 - 1802292 --AAGAGAGCCGCUCCGCUGCUGUCGUAUCCGGCAGCGCAGCUACGCGGCUUAGCCUUCGACG-AAAUAUAAUUAAGAAUCACAAAACAGA--- --....(((((((...((((((((((....)))))..)))))...)))))))...........-...........................--- ( -26.00, z-score = -1.10, R) >droSim1.chr3h_random 100612 79 + 1452968 --AUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGACUUAGGCUAAGAAA-CCAUUGUAUUUAGAUCGC------------ --....(((((((.(((((((.(((...))).))))))).))...........((........-))..........)).)))------------ ( -26.70, z-score = -0.56, R) >droPer1.super_51 391943 88 + 524598 -AAUGCG-GCCGCUGCGCUGACGGCUUUUCUGGCAGCGCUGCUGCUCGGCCUUAGCCAAGUAA-ACUUUAAAUUAAAAGAUAUGAAAGCAG--- -..((((-((.((.((((((.(((.....))).)))))).)).))).(((....)))......-.((((......))))........))).--- ( -27.10, z-score = -0.11, R) >dp4.Unknown_group_24 89643 88 + 109830 -AAUGCG-GCCGCUGCGCUGACGGCUUUUCUGGCAGCGCUGCUGCUCGGCCUUAGCCAAGUAA-ACUUUAAAUUAAAAGAUAUGAAAGCAG--- -..((((-((.((.((((((.(((.....))).)))))).)).))).(((....)))......-.((((......))))........))).--- ( -27.10, z-score = -0.11, R) >droVir3.scaffold_13036 610849 85 - 1135500 GUAUGAA-GCCGCUCCGCCGCGGGGGCGUUCGGCAGUGCGGCUGUGCGGGCUUGGCCAUAUGG-AGAUAGAGGGUAAGACACUCUGA------- (((((((-(((((.(.((((((..(.(....).)..)))))).).)).)))))...)))))..-...((((((......).))))).------- ( -30.80, z-score = 0.60, R) >consensus __AUGAG_GCCGCUCCGCUGCUGGCGUUUCCGGCAGCGCAGCUGCGCGGCCUAGGCUAAGUAA_AAAUAAAAUUAAAAACUAUAAAA_______ ........(((((..(((((((((.....)))))))))..)).....)))............................................ (-12.63 = -12.81 + 0.19)

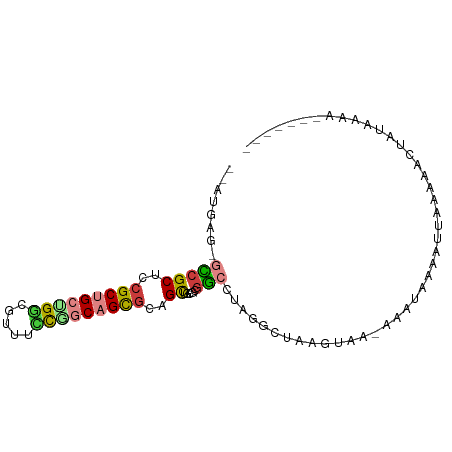

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:54 2011