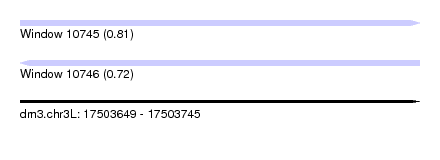
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,503,649 – 17,503,745 |
| Length | 96 |
| Max. P | 0.810054 |

| Location | 17,503,649 – 17,503,745 |
|---|---|
| Length | 96 |
| Sequences | 13 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 61.01 |
| Shannon entropy | 0.85380 |
| G+C content | 0.45289 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -10.10 |
| Energy contribution | -10.66 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.46 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17503649 96 + 24543557 ------GGCAAACAGAUGUUACACUAACGAUAA--CAAAACCCUUAAUUCCCCG-CAGAUAAGGCCAUGGCCAUGGGUGUGAUACAGUCGGCCAUCGGCCUGUUC ------....(((...(((((..(....).)))--)).................-......(((((((((((..((.((.....)).)))))))).)))))))). ( -23.70, z-score = -0.01, R) >droEre2.scaffold_4784 9418660 94 - 25762168 ------CAAAAACAGAUGUUACACUAACGAUAA--CAAAACCC-AAAU-CCCCA-CAGAUAAGGCCAUGGCCAUGGGUGUGAUACAGUCGGCCAUCGGCCUGUUC ------....(((...(((((..(....).)))--))......-....-.....-......(((((((((((..((.((.....)).)))))))).)))))))). ( -23.70, z-score = -1.11, R) >droYak2.chr3L 7635829 94 - 24197627 ------AGAAAACAUAUGUUACACCAACGAUAA--CCAAACCU-AAAU-CCCCA-CAGAUAAGGCCAUGGCCAUGGGUGUGAUACAGUCGGCCAUCGGCCUGUUC ------....((((..(((((((((........--........-..((-(....-..)))..(((....)))...))))))))).....((((...)))))))). ( -23.90, z-score = -0.99, R) >droSec1.super_0 9555851 95 + 21120651 ------GGCAAACAGAUGUUACACUAACGAUAA--CAAAACCCUAAAU-CCCCG-CAGAUAAGGCCAUGGCCAUGGGUGUGAUACAGUCGGCCAUCGGCCUGUUC ------....(((...(((((..(....).)))--))...........-.....-......(((((((((((..((.((.....)).)))))))).)))))))). ( -23.70, z-score = -0.13, R) >droSim1.chr3L 16843819 95 + 22553184 ------GGCAAACAGAUGUUACACUAACGAUAA--CAAAACCCUAAAU-CCCCG-CAGAUAAGGCCAUGGCCAUGGGUGUGAUACAGUCGGCCAUCGGCCUGUUC ------....(((...(((((..(....).)))--))...........-.....-......(((((((((((..((.((.....)).)))))))).)))))))). ( -23.70, z-score = -0.13, R) >droWil1.scaffold_181009 189372 90 - 3585778 ------------AAUCCGUUGA-UAUCUAAUAUCACAACAAUU--CUCAUUAAAACAGAUAAAGCCAUGGCCAUGGGUGUCAUACAGUCGGCCAUUGGCCUAUUU ------------.....((.((-(((...))))))).......--..................(((((((((..((.((.....)).))))))).))))...... ( -18.90, z-score = -1.14, R) >dp4.chrXR_group6 4185977 98 - 13314419 ------UACCAGUAUAUCUAGAUCAAUCGGUAAACUAAAUACUCUUAUGACUUG-CAGACAAGGCCAUGGCCAUGGGUGUGAUACAGUCUGCCAUUGGCUUAUUC ------............(((..((((.((((.(((...(((.((((((.((((-....))))((....)))))))).)))....))).))))))))..)))... ( -21.60, z-score = 0.09, R) >droPer1.super_35 594945 98 + 973955 ------UACCAGUAUAUCUAGAUCAAUCGGUAAACUAUAUACUCUUAUGACUUG-CAGACAAGGCCAUGGCCAUGGGUGUGAUACAGUCUGCCAUUGGCUUAUUC ------............(((..((((.((((.(((.(((((.((((((.((((-....))))((....)))))))).)).))).))).))))))))..)))... ( -22.90, z-score = -0.11, R) >droAna3.scaffold_13337 14213522 89 + 23293914 ---------------GGAAGGGAAUGUCAAAUAAUUUGGAAUAUUUUUCACUUU-CAGACAAGGCCAUGGCCAUGGGAGUGAUACAGUCGGCCAUCGGCUUGUUC ---------------(((((((((..((((.....))))......)))).))))-).((((((.((((((((.((.........))...)))))).)))))))). ( -24.70, z-score = -1.41, R) >droVir3.scaffold_13049 8172430 84 + 25233164 ---------------GCAGAGAAUUACCAACUG-----AUAUACACUU-UCAUACCAGAUAAAGCUAUGGCUAUGGGCGUCAUUCAGUCGGCCAUCGGCCUGUUU ---------------...............(((-----.(((......-..))).)))...((((...((((((((.((.(.....).)).)))).)))).)))) ( -17.90, z-score = -0.10, R) >droMoj3.scaffold_6680 11834729 95 + 24764193 ----------ACCAGGCGUAGAAUCUCAAAUCUCAUGAACUUUACUUUACCCAACUAGAUAAAGCCAUGGCCAUGGGCGUCAUCCAGUCAGCCAUCGGCCUGUUU ----------..((((((((((...(((.......)))..)))))...................(.(((((..((((.....))))....))))).))))))... ( -20.10, z-score = -0.23, R) >droGri2.scaffold_15110 7201643 89 + 24565398 -------------ACCCAUUAAAUAUACCCUAA--UUAAAAUGCACUCUUCAAA-UAGAUAAGGCCAUGGCAAUGGGUGUUAUACAGUCGGCCAUUGGUCUAUUU -------------....................--...........(((.....-.)))..((((((((((...((.((.....)).)).)))).)))))).... ( -16.90, z-score = 0.05, R) >anoGam1.chr2L 13961481 105 - 48795086 UGGGAGGCUGUAUAAAUCAAAAGCGUACUCCUUAUUUGUAGAUGUGUUGAACCACGCGAUAAAGCCAUCUCGAUGGGAUUCUCGAUGGCGAUCAACACACUGUUC .((((((((............)))...)))))......(((.((((((((..(....).....((((((..((......))..))))))..)))))))))))... ( -26.70, z-score = -0.70, R) >consensus __________AACAGAUGUUAAACUAACGAUAA__CAAAAACUCAAAU_CCCCA_CAGAUAAGGCCAUGGCCAUGGGUGUGAUACAGUCGGCCAUCGGCCUGUUC ...............................................................((((((((..(((.((.....)).)))))))).)))...... (-10.10 = -10.66 + 0.56)

| Location | 17,503,649 – 17,503,745 |
|---|---|
| Length | 96 |
| Sequences | 13 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 61.01 |
| Shannon entropy | 0.85380 |
| G+C content | 0.45289 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -10.12 |
| Energy contribution | -10.33 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.54 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17503649 96 - 24543557 GAACAGGCCGAUGGCCGACUGUAUCACACCCAUGGCCAUGGCCUUAUCUG-CGGGGAAUUAAGGGUUUUG--UUAUCGUUAGUGUAACAUCUGUUUGCC------ ((..(((((.(((((((...((.....))...))))))))))))..)).(-((((..(((((.(((....--..))).)))))......))))).....------ ( -26.70, z-score = 0.14, R) >droEre2.scaffold_4784 9418660 94 + 25762168 GAACAGGCCGAUGGCCGACUGUAUCACACCCAUGGCCAUGGCCUUAUCUG-UGGGG-AUUU-GGGUUUUG--UUAUCGUUAGUGUAACAUCUGUUUUUG------ (((((((..((((((.((((..(((...((((((((....)))......)-)))))-))..-.))))..)--)))))((((...)))).)))))))...------ ( -24.20, z-score = 0.39, R) >droYak2.chr3L 7635829 94 + 24197627 GAACAGGCCGAUGGCCGACUGUAUCACACCCAUGGCCAUGGCCUUAUCUG-UGGGG-AUUU-AGGUUUGG--UUAUCGUUGGUGUAACAUAUGUUUUCU------ (((((((((...)))).........(((((.(((((((.(((((.((((.-...))-))..-))))))))--))))....)))))......)))))...------ ( -27.90, z-score = -0.37, R) >droSec1.super_0 9555851 95 - 21120651 GAACAGGCCGAUGGCCGACUGUAUCACACCCAUGGCCAUGGCCUUAUCUG-CGGGG-AUUUAGGGUUUUG--UUAUCGUUAGUGUAACAUCUGUUUGCC------ ((..(((((.(((((((...((.....))...))))))))))))..))..-..((.-(..((((....((--(((((....).))))))))))..).))------ ( -25.30, z-score = 0.56, R) >droSim1.chr3L 16843819 95 - 22553184 GAACAGGCCGAUGGCCGACUGUAUCACACCCAUGGCCAUGGCCUUAUCUG-CGGGG-AUUUAGGGUUUUG--UUAUCGUUAGUGUAACAUCUGUUUGCC------ ((..(((((.(((((((...((.....))...))))))))))))..))..-..((.-(..((((....((--(((((....).))))))))))..).))------ ( -25.30, z-score = 0.56, R) >droWil1.scaffold_181009 189372 90 + 3585778 AAAUAGGCCAAUGGCCGACUGUAUGACACCCAUGGCCAUGGCUUUAUCUGUUUUAAUGAG--AAUUGUUGUGAUAUUAGAUA-UCAACGGAUU------------ ....((((((.((((((...((.....))...)))))))))))).(((((((.((((...--.))))....(((((...)))-))))))))).------------ ( -22.00, z-score = -1.17, R) >dp4.chrXR_group6 4185977 98 + 13314419 GAAUAAGCCAAUGGCAGACUGUAUCACACCCAUGGCCAUGGCCUUGUCUG-CAAGUCAUAAGAGUAUUUAGUUUACCGAUUGAUCUAGAUAUACUGGUA------ ......(((....((((((((.....)).....(((....)))..)))))-)........((.((((((((.(((.....))).)))))))).))))).------ ( -24.10, z-score = -0.76, R) >droPer1.super_35 594945 98 - 973955 GAAUAAGCCAAUGGCAGACUGUAUCACACCCAUGGCCAUGGCCUUGUCUG-CAAGUCAUAAGAGUAUAUAGUUUACCGAUUGAUCUAGAUAUACUGGUA------ ......(((....((((((((.....)).....(((....)))..)))))-)..........(((((((((.(((.....))).)))..))))))))).------ ( -21.70, z-score = 0.10, R) >droAna3.scaffold_13337 14213522 89 - 23293914 GAACAAGCCGAUGGCCGACUGUAUCACUCCCAUGGCCAUGGCCUUGUCUG-AAAGUGAAAAAUAUUCCAAAUUAUUUGACAUUCCCUUCC--------------- ..(((((((.(((((((...(.....).....))))))))).)))))...-..((((..(((((........)))))..)))).......--------------- ( -18.60, z-score = -1.29, R) >droVir3.scaffold_13049 8172430 84 - 25233164 AAACAGGCCGAUGGCCGACUGAAUGACGCCCAUAGCCAUAGCUUUAUCUGGUAUGA-AAGUGUAUAU-----CAGUUGGUAAUUCUCUGC--------------- ...((((..((..(((((((((...((((.((((.(((.(......).))))))).-..))))...)-----))))))))...)))))).--------------- ( -24.50, z-score = -1.89, R) >droMoj3.scaffold_6680 11834729 95 - 24764193 AAACAGGCCGAUGGCUGACUGGAUGACGCCCAUGGCCAUGGCUUUAUCUAGUUGGGUAAAGUAAAGUUCAUGAGAUUUGAGAUUCUACGCCUGGU---------- ...(((((...((.(..(((((((((.(((.((....))))).)))))))))..).))..(((...((((.......))))....))))))))..---------- ( -31.80, z-score = -2.24, R) >droGri2.scaffold_15110 7201643 89 - 24565398 AAAUAGACCAAUGGCCGACUGUAUAACACCCAUUGCCAUGGCCUUAUCUA-UUUGAAGAGUGCAUUUUAA--UUAGGGUAUAUUUAAUGGGU------------- ...........................((((((((.....((((((....-.((((((......))))))--.)))))).....))))))))------------- ( -15.80, z-score = 0.92, R) >anoGam1.chr2L 13961481 105 + 48795086 GAACAGUGUGUUGAUCGCCAUCGAGAAUCCCAUCGAGAUGGCUUUAUCGCGUGGUUCAACACAUCUACAAAUAAGGAGUACGCUUUUGAUUUAUACAGCCUCCCA .....(((((((((..((((((((.((..((((....))))..)).))).))))))))))))))..........((((...(((.(........).))))))).. ( -28.40, z-score = -2.03, R) >consensus GAACAGGCCGAUGGCCGACUGUAUCACACCCAUGGCCAUGGCCUUAUCUG_UGGGG_AUUUAAGUUUUUA__UUAUCGUUAGUUUAACAUCUGUU__________ ....((((((.((((...................))))))))))............................................................. (-10.12 = -10.33 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:53 2011