
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,492,622 – 17,492,720 |
| Length | 98 |
| Max. P | 0.983838 |

| Location | 17,492,622 – 17,492,720 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 72.66 |
| Shannon entropy | 0.45693 |
| G+C content | 0.56473 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -15.04 |
| Energy contribution | -15.96 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.822303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
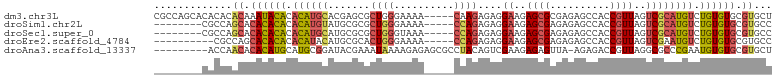
>dm3.chr3L 17492622 98 + 24543557 CGCCAGCACACACACAAAUACACACAUGCACGAGCGCUGGGAAAA-----CAAGAGAGGAAGAGCGCGAGAGCCACCGUUAGUCGCAUGUCUGUGUGCGUGCU ....(((((.((((((.......((((((....(((((.......-----............)))))((.(((....)))..)))))))).)))))).))))) ( -29.51, z-score = -0.37, R) >droSim1.chr2L 658062 90 + 22036055 --------CGCCAGCACACACACACAUGUAUGCGCGCUGGGAAAA-----CCAGAGAGGAAGAGCGAGAGAGCCACCGUUAGUCGCAUGUCUGUGUGCGUGCC --------.....((((...((((((..(((((((.((((.....-----))))...((..(..(....)..)..))....).))))))..)))))).)))). ( -32.00, z-score = -1.58, R) >droSec1.super_0 9545149 90 + 21120651 --------CGCCAGCACACACACACAUGCAUGCGCGCUGGGUAAA-----CCAGAGAGGAAGAGCGAGAGAGCCACCGUUAGUCGCAUGUCUGUGUGCGUGCC --------.....((((.((((((...((((((((.((((.....-----))))...((..(..(....)..)..))....).))))))).)))))).)))). ( -35.60, z-score = -2.19, R) >droEre2.scaffold_4784 9405319 88 - 25762168 ----------CGCCAGCACACACACAUACAUGCGCACUGGGAAAA-----CCAGAGAGGAAGAGCGAGAGAGCCACCGUUAGUCGAAUGUCUGUGUGCGUGCC ----------.....((((.((((((.((((.(((.((((.....-----))))...((..(..(....)..)..))....).)).)))).)))))).)))). ( -31.40, z-score = -2.79, R) >droAna3.scaffold_13337 14202845 93 + 23293914 ---------ACCAACACACAUGCAUGCGGAUACGAAAUAAAAGAGAGCGCCUACAGUCGAAGAGAGUUA-AGAGACCGUUAGGCGCCCGAAUGUGUGCGUGCU ---------.....(((.(((((((.(((...(.........)...((((((((.(((...........-...))).).)))))))))).))))))).))).. ( -28.14, z-score = -2.89, R) >consensus ________CACCAACACACACACACAUGCAUGCGCGCUGGGAAAA_____CCAGAGAGGAAGAGCGAGAGAGCCACCGUUAGUCGCAUGUCUGUGUGCGUGCC .............((.((((((.((((((.......((((..........))))....((..((((..........))))..)))))))).)))))).))... (-15.04 = -15.96 + 0.92)

| Location | 17,492,622 – 17,492,720 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 72.66 |
| Shannon entropy | 0.45693 |
| G+C content | 0.56473 |
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -18.72 |
| Energy contribution | -20.64 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
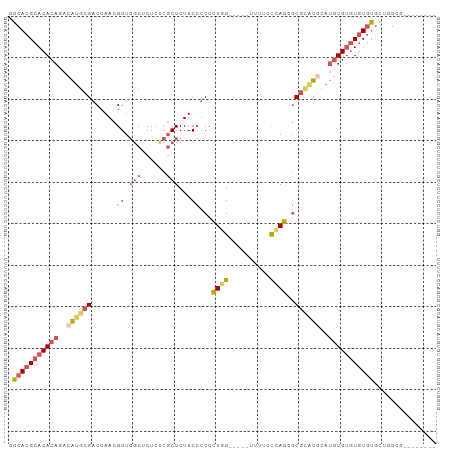
>dm3.chr3L 17492622 98 - 24543557 AGCACGCACACAGACAUGCGACUAACGGUGGCUCUCGCGCUCUUCCUCUCUUG-----UUUUCCCAGCGCUCGUGCAUGUGUGUAUUUGUGUGUGUGCUGGCG ((((((((((((.(((((((.(....)...((.(..(((((......(....)-----.......)))))..).)).)))))))...)))))))))))).... ( -34.92, z-score = -1.36, R) >droSim1.chr2L 658062 90 - 22036055 GGCACGCACACAGACAUGCGACUAACGGUGGCUCUCUCGCUCUUCCUCUCUGG-----UUUUCCCAGCGCGCAUACAUGUGUGUGUGUGCUGGCG-------- ((((((((((((.(.(((((......((.(((......)))...))...((((-----.....))))..)))))...).))))))))))))....-------- ( -33.60, z-score = -1.79, R) >droSec1.super_0 9545149 90 - 21120651 GGCACGCACACAGACAUGCGACUAACGGUGGCUCUCUCGCUCUUCCUCUCUGG-----UUUACCCAGCGCGCAUGCAUGUGUGUGUGUGCUGGCG-------- ((((((((((((.(((((((......((.(((......)))...))...((((-----.....))))..))))))..).))))))))))))....-------- ( -36.50, z-score = -1.99, R) >droEre2.scaffold_4784 9405319 88 + 25762168 GGCACGCACACAGACAUUCGACUAACGGUGGCUCUCUCGCUCUUCCUCUCUGG-----UUUUCCCAGUGCGCAUGUAUGUGUGUGUGCUGGCG---------- ((((((((((((.((((.((......((.(((......)))...)).(.((((-----.....)))).))).)))).))))))))))))....---------- ( -33.60, z-score = -2.70, R) >droAna3.scaffold_13337 14202845 93 - 23293914 AGCACGCACACAUUCGGGCGCCUAACGGUCUCU-UAACUCUCUUCGACUGUAGGCGCUCUCUUUUAUUUCGUAUCCGCAUGCAUGUGUGUUGGU--------- ..((.((((((((..(((((((((.(((((...-...........))))))))))))))...........((((....))))))))))))))..--------- ( -33.14, z-score = -4.32, R) >consensus GGCACGCACACAGACAUGCGACUAACGGUGGCUCUCUCGCUCUUCCUCUCUGG_____UUUUCCCAGCGCGCAUGCAUGUGUGUGUGUGCUGGCG________ .(((((((((((..((((((......((.(((......)))...))...((((..........))))..))))))..)))))))))))............... (-18.72 = -20.64 + 1.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:52 2011