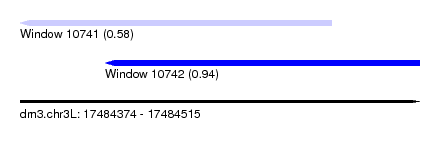
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,484,374 – 17,484,515 |
| Length | 141 |
| Max. P | 0.939104 |

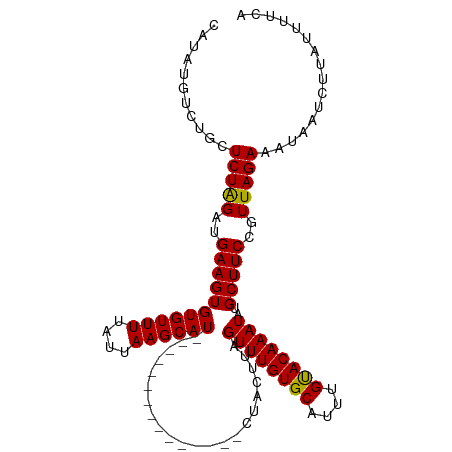
| Location | 17,484,374 – 17,484,484 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.00 |
| Shannon entropy | 0.26351 |
| G+C content | 0.31545 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -15.62 |
| Energy contribution | -17.07 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.578905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17484374 110 - 24543557 UUAAGCAUUGUCUGUUGUUCAUCCUACUUAGUUUGUGCAUUUGCACAAAUAUGCUUCCGUUAGAAAUAAUCUUGUAAUCAUACGAUGAGUA---CUAUCCCUU--AUUUCAGUUC ..((((((.(..((.....))..)......((((((((....))))))))))))))......(((((((....(((.((((...)))).))---)......))--)))))..... ( -20.00, z-score = -1.68, R) >droSec1.super_0 9537019 100 - 21120651 --------------UUAAGCAU-CUACUUAGUUUGUGCAUUUGUACAAAUAUGCUUCCGUUAGAAAUAAUCUUAUUUUCAGACGAUGAGUAGGGAUAGUACUUAUAUUUCAGUUC --------------.......(-(((((((((((((((....)))))))).......((((.((((.........)))).)))).))))))))...................... ( -20.90, z-score = -1.57, R) >droSim1.chr3L 16825346 100 - 22553184 --------------UUAAGCAU-CUACUUAGUUUGUGCAUUUGUACAAAUAUGCUUCCGUUAGAAAUAAUCUUAUUUUAAGACGAUGAGUAGGGAUAGUACUUAUAUUUCAGUUC --------------.......(-(((((((((((((((....)))))))).......((((.((((((....))))))..)))).))))))))...................... ( -19.40, z-score = -1.05, R) >consensus ______________UUAAGCAU_CUACUUAGUUUGUGCAUUUGUACAAAUAUGCUUCCGUUAGAAAUAAUCUUAUUUUCAGACGAUGAGUAGGGAUAGUACUUAUAUUUCAGUUC .......................(((((((((((((((....)))))))).......((((.((((.........)))).)))).)))))))....................... (-15.62 = -17.07 + 1.45)

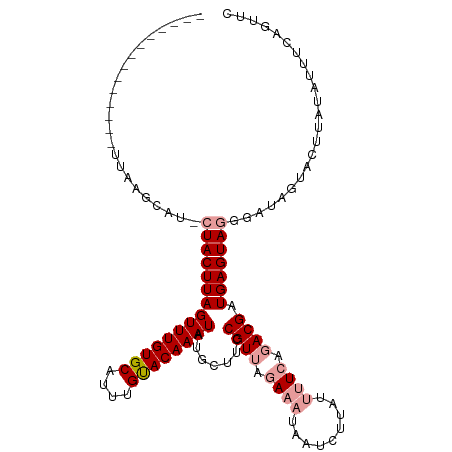
| Location | 17,484,404 – 17,484,515 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 85.53 |
| Shannon entropy | 0.19028 |
| G+C content | 0.31898 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -19.74 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.939104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17484404 111 - 24543557 CAUAUGUCUGCUCUGGAUGAAGUGUGUUUUAUUAAGCAUUGUCUGUUGUUCAUCCUACUUAGUUUGUGCAUUUGCACAAAUAUGCUUCCGUUAGAAAUAAUCUUGUAAUCA ..............((((((((.((((((....))))))...)).....))))))......((((((((....)))))))).(((....((((....))))...))).... ( -22.50, z-score = -1.08, R) >droSec1.super_0 9537054 96 - 21120651 CAUGUGUCUGCUCUAGAUGAAGUGUGUUUUAUUAAGCAU---------------CUACUUAGUUUGUGCAUUUGUACAAAUAUGCUUCCGUUAGAAAUAAUCUUAUUUUCA ...........(((((..(((((((((((.((((((...---------------...))))))..((((....)))))))))))))))..)))))................ ( -18.90, z-score = -1.35, R) >droSim1.chr3L 16825381 96 - 22553184 CAUAUGUCUGCUCUAGCUGAAGUGUGUUUUAUUAAGCAU---------------CUACUUAGUUUGUGCAUUUGUACAAAUAUGCUUCCGUUAGAAAUAAUCUUAUUUUAA ...........((((((.(((((((((((.((((((...---------------...))))))..((((....))))))))))))))).))))))................ ( -22.80, z-score = -2.95, R) >consensus CAUAUGUCUGCUCUAGAUGAAGUGUGUUUUAUUAAGCAU_______________CUACUUAGUUUGUGCAUUUGUACAAAUAUGCUUCCGUUAGAAAUAAUCUUAUUUUCA ...........(((((..(((((((((((....))))))......................((((((((....))))))))..)))))..)))))................ (-19.74 = -19.30 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:50 2011