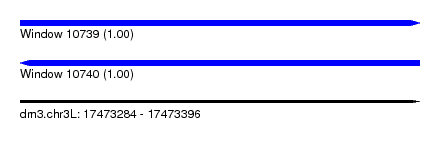
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,473,284 – 17,473,396 |
| Length | 112 |
| Max. P | 0.999402 |

| Location | 17,473,284 – 17,473,396 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.33 |
| Shannon entropy | 0.20998 |
| G+C content | 0.37606 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -23.85 |
| Energy contribution | -24.85 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -4.61 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17473284 112 + 24543557 UGCUAAUUUAAGCUGACCACAAAACCCAGUGAAACCACAAUGUACACACAAGAAUAC--------GAUUCACAACAUAUUCCUGUACCUACUUGUGUUUUCAUUGGACUUCUUGGUCGAA .(((......)))((((((..((..(((((((((.(((((.(((...(((.(((((.--------(........).))))).)))...)))))))).)))))))))..))..)))))).. ( -32.40, z-score = -5.04, R) >droSim1.chr3L 16814122 111 + 22553184 UGCUAAUUUAAGCUGACCACAAAACCCAAUGGAACCACA-UGUACACACAAGAAUAU--------GAUUCACAACCUAUUCCUGUACCUACUUGUGUUUUCAUUGGCGUUCUUGGUCGAA .(((......)))((((((...((((((((((((.((((-.(((...(((.(((((.--------(........).))))).)))...))).)))).))))))))).)))..)))))).. ( -33.30, z-score = -4.90, R) >droSec1.super_0 9526184 112 + 21120651 UGCUAAUUUAAGCUGACCACAAAACCCAAUGGAACCACAAUGUACACACAAGAAUAU--------GAUUCACAACCUAUUCCUGUACCUACUUGUGUUUUCAUUGGCGUUCUUGGUCGAA .(((......)))((((((...((((((((((((.(((((.(((...(((.(((((.--------(........).))))).)))...)))))))).))))))))).)))..)))))).. ( -33.60, z-score = -4.99, R) >droYak2.chr3L 7604678 112 - 24197627 UGAUAAUUUAAACUGACCAGAAAACCCAUUGAAAACACAAUGUACACACAGCAAUAC--------GAUUCACAACAUAUUCCUGUAGUUACUUGUGUUUCCAUUAGAGUUCCUGGUCGAA .............(((((((.((......((.((((((((.(((.((((((.((((.--------(........).)))).)))).))))))))))))).))......)).))))))).. ( -26.10, z-score = -3.02, R) >droEre2.scaffold_4784 9386795 120 - 25762168 UGAUAAUUUAAACUGACCAGAAAACCCAUUGAAAACACAAUGUACAUACAGGAAUACAUGGGUACGAAUCACAACAUAUUCCUGUAGUUACCUGUGUUUUUAUUGGAGUUCUUGGUCGAA .............(((((((((...(((.((((((((((..(((..((((((((((..(((((....))).))...))))))))))..))).)))))))))).)))..))).)))))).. ( -40.00, z-score = -5.12, R) >consensus UGCUAAUUUAAGCUGACCACAAAACCCAAUGAAACCACAAUGUACACACAAGAAUAC________GAUUCACAACAUAUUCCUGUACCUACUUGUGUUUUCAUUGGAGUUCUUGGUCGAA .............((((((..((..(((((((((.(((((.(((...(((.(((((....................))))).)))...)))))))).)))))))))..))..)))))).. (-23.85 = -24.85 + 1.00)

| Location | 17,473,284 – 17,473,396 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.33 |
| Shannon entropy | 0.20998 |
| G+C content | 0.37606 |
| Mean single sequence MFE | -43.74 |
| Consensus MFE | -32.85 |
| Energy contribution | -33.53 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.15 |
| Mean z-score | -6.10 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17473284 112 - 24543557 UUCGACCAAGAAGUCCAAUGAAAACACAAGUAGGUACAGGAAUAUGUUGUGAAUC--------GUAUUCUUGUGUGUACAUUGUGGUUUCACUGGGUUUUGUGGUCAGCUUAAAUUAGCA ...(((((.(((..(((.(((((.((((((((..((((((((((((........)--------)))))))))))..))).))))).))))).)))..))).))))).(((......))). ( -47.10, z-score = -7.14, R) >droSim1.chr3L 16814122 111 - 22553184 UUCGACCAAGAACGCCAAUGAAAACACAAGUAGGUACAGGAAUAGGUUGUGAAUC--------AUAUUCUUGUGUGUACA-UGUGGUUCCAUUGGGUUUUGUGGUCAGCUUAAAUUAGCA ...((((((((((.((((((....((((.(((..(((((((((((((.....)))--------.))))))))))..))).-))))....))))))))))).))))).(((......))). ( -42.40, z-score = -5.45, R) >droSec1.super_0 9526184 112 - 21120651 UUCGACCAAGAACGCCAAUGAAAACACAAGUAGGUACAGGAAUAGGUUGUGAAUC--------AUAUUCUUGUGUGUACAUUGUGGUUCCAUUGGGUUUUGUGGUCAGCUUAAAUUAGCA ...((((((((((.((((((....((((((((..(((((((((((((.....)))--------.))))))))))..))).)))))....))))))))))).))))).(((......))). ( -42.70, z-score = -5.49, R) >droYak2.chr3L 7604678 112 + 24197627 UUCGACCAGGAACUCUAAUGGAAACACAAGUAACUACAGGAAUAUGUUGUGAAUC--------GUAUUGCUGUGUGUACAUUGUGUUUUCAAUGGGUUUUCUGGUCAGUUUAAAUUAUCA ...(((((((((..(((.((((((((((((((..(((((.((((((........)--------))))).)))))..))).))))))))))).)))..))))))))).............. ( -45.30, z-score = -7.02, R) >droEre2.scaffold_4784 9386795 120 + 25762168 UUCGACCAAGAACUCCAAUAAAAACACAGGUAACUACAGGAAUAUGUUGUGAUUCGUACCCAUGUAUUCCUGUAUGUACAUUGUGUUUUCAAUGGGUUUUCUGGUCAGUUUAAAUUAUCA ...((((((((((.(((.(.((((((((((((..(((((((((((((.(((.....)))..)))))))))))))..))).))))))))).).)))))))).))))).............. ( -41.20, z-score = -5.41, R) >consensus UUCGACCAAGAACUCCAAUGAAAACACAAGUAGGUACAGGAAUAUGUUGUGAAUC________GUAUUCUUGUGUGUACAUUGUGGUUUCAAUGGGUUUUGUGGUCAGCUUAAAUUAGCA ...((((((((((.(((.(((((.((((((((..((((((((((....................))))))))))..))).))))).))))).)))))))).))))).(((......))). (-32.85 = -33.53 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:49 2011