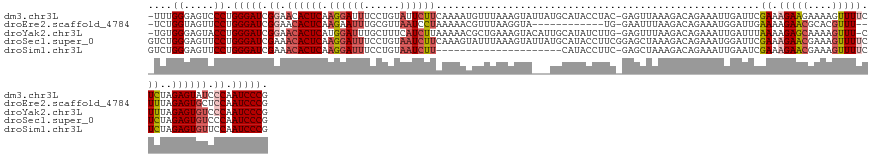
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,446,551 – 17,446,689 |
| Length | 138 |
| Max. P | 0.999530 |

| Location | 17,446,551 – 17,446,689 |
|---|---|
| Length | 138 |
| Sequences | 5 |
| Columns | 140 |
| Reading direction | forward |
| Mean pairwise identity | 76.53 |
| Shannon entropy | 0.41669 |
| G+C content | 0.39794 |
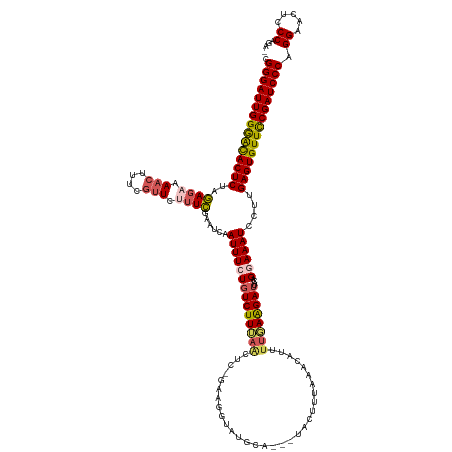
| Mean single sequence MFE | -40.81 |
| Consensus MFE | -30.88 |
| Energy contribution | -31.20 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.68 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.98 |
| SVM RNA-class probability | 0.999530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
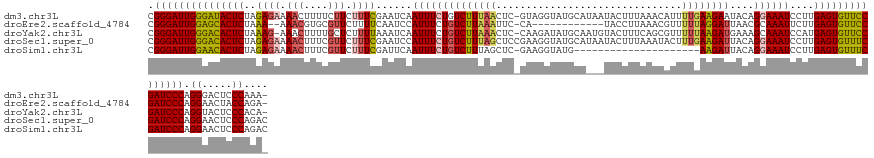
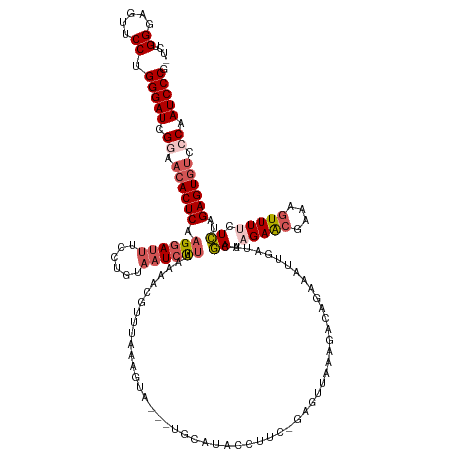
>dm3.chr3L 17446551 138 + 24543557 CGGGAUUGGGAUACUCUAGAGAAAACUUUUCUUCUUUCGAAUCAAUUUCUGUCUUUAACUC-GUAGGUAUGCAUAAUACUUUAAACAUUUUGAAGAAUACAGGAAAUCCUUGAGUGUUCCGAUCCCAGGGACUCCCAAA- .(((((((((((((((.((((((........))))))........(((((((((((((...-((((((((.....))))))...))...)))))))...))))))......))))))))))))))).(((...)))...- ( -43.20, z-score = -3.81, R) >droEre2.scaffold_4784 9361095 124 - 25762168 CGGGAUUGGAGCACUCUAAA--AAACGUGCGUUCUUUUCAAUCCAUUUCUGUCUUAAAUUC-CA------------UACCUUAAACGUUUUUAGGAUUAACGCAAAUUCUUGAGUGUUCCGAUCCCAGGAACUACCAGA- .(((((((((((((((....--.....((((((.................((((((((..(-..------------..........)..)))))))).)))))).......))))))))))))))).((.....))...- ( -38.16, z-score = -4.62, R) >droYak2.chr3L 7578039 137 - 24197627 CGGGAUUGGGACACUCUAAAG-AAACUUUUGCUCUUUUAAAUCAAUUUCUGUCUUAAACUC-CAAGAUAUGCAAUGUACUUUCAGCGUUUUUAAGAUGAAAGCAAAUCCAUGAGUGUUCCGAUCCCAGGUACUCCCACA- .(((((((((((((((....(-(....((((((.(((((((........((((((......-.))))))...(((((.......))))))))))))....))))))))...))))))))))))))).((.....))...- ( -37.70, z-score = -3.32, R) >droSec1.super_0 9499237 140 + 21120651 CGGGAUUGGGACACUCUAGAGAAAACUUUCGUUCUUUCGAAUCCAUUUCUGUCUUUAGCUCCGAAGGUAUGCAUAAUACUUUAAAUACUUUGAAGAUUACAGGAAAUCCUUGAGUGUUUCGAUCCCAGGAACUCCCAGAC .(((((((..((((((.((.((.....((((......))))....(((((((((((((.....(((((((.....))))))).......)))))))...)))))).)))).))))))..))))))).((.....)).... ( -44.50, z-score = -3.54, R) >droSim1.chr3L 16786903 118 + 22553184 CGGGAUUGGAACACUCUAGAGAAAACUUUCGUUCUUUCGAUUCAAUUUCUGUCUUUAGCUC-GAAGGUAUG---------------------AAGAUUACAGGAAAUCCUUGAGUGUUUCGAUCCCAGGAACUCCCAGAC .(((((((((((((((.((.((......(((......))).....((((((((((((...(-....)..))---------------------))))...)))))).)))).))))))))))))))).((.....)).... ( -40.50, z-score = -3.14, R) >consensus CGGGAUUGGGACACUCUAGAGAAAACUUUCGUUCUUUCGAAUCAAUUUCUGUCUUUAACUC_GAAGGUAUGCA___UACUUUAAACAUUUUGAAGAUUACAGGAAAUCCUUGAGUGUUCCGAUCCCAGGAACUCCCAGA_ .(((((((((((((((..((((.(((....))).))))......((((((((((((((...............................))))))))....))))))....))))))))))))))).((.....)).... (-30.88 = -31.20 + 0.32)
| Location | 17,446,551 – 17,446,689 |
|---|---|
| Length | 138 |
| Sequences | 5 |
| Columns | 140 |
| Reading direction | reverse |
| Mean pairwise identity | 76.53 |
| Shannon entropy | 0.41669 |
| G+C content | 0.39794 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -21.10 |
| Energy contribution | -22.50 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17446551 138 - 24543557 -UUUGGGAGUCCCUGGGAUCGGAACACUCAAGGAUUUCCUGUAUUCUUCAAAAUGUUUAAAGUAUUAUGCAUACCUAC-GAGUUAAAGACAGAAAUUGAUUCGAAAGAAGAAAAGUUUUCUCUAGAGUAUCCCAAUCCCG -...(((...))).(((((.((...((((.(((....)))...((((((...((((.(((....))).)))).....(-(((((((.........))))))))...))))))............))))...)).))))). ( -33.30, z-score = -1.25, R) >droEre2.scaffold_4784 9361095 124 + 25762168 -UCUGGUAGUUCCUGGGAUCGGAACACUCAAGAAUUUGCGUUAAUCCUAAAAACGUUUAAGGUA------------UG-GAAUUUAAGACAGAAAUGGAUUGAAAAGAACGCACGUUU--UUUAGAGUGCUCCAAUCCCG -...((.....)).(((((.(((.(((((((((((.((((((..(((......(......)...------------.)-)).(((((..((....))..)))))...)))))).))))--))..))))).))).))))). ( -38.40, z-score = -3.55, R) >droYak2.chr3L 7578039 137 + 24197627 -UGUGGGAGUACCUGGGAUCGGAACACUCAUGGAUUUGCUUUCAUCUUAAAAACGCUGAAAGUACAUUGCAUAUCUUG-GAGUUUAAGACAGAAAUUGAUUUAAAAGAGCAAAAGUUU-CUUUAGAGUGUCCCAAUCCCG -...((.....)).(((((.((.((((((..(((..((((((((.(........).))))))))..((((...((((.-.((.((((........)))).))..)))))))).....)-))...)))))).)).))))). ( -35.50, z-score = -1.59, R) >droSec1.super_0 9499237 140 - 21120651 GUCUGGGAGUUCCUGGGAUCGAAACACUCAAGGAUUUCCUGUAAUCUUCAAAGUAUUUAAAGUAUUAUGCAUACCUUCGGAGCUAAAGACAGAAAUGGAUUCGAAAGAACGAAAGUUUUCUCUAGAGUGUCCCAAUCCCG ....(((....)))(((((.(..((((((..((((((.((((...((((((.((((.((........)).)))).)).))))......))))....))))))((.(((((....))))).))..))))))..).))))). ( -34.80, z-score = -0.81, R) >droSim1.chr3L 16786903 118 - 22553184 GUCUGGGAGUUCCUGGGAUCGAAACACUCAAGGAUUUCCUGUAAUCUU---------------------CAUACCUUC-GAGCUAAAGACAGAAAUUGAAUCGAAAGAACGAAAGUUUUCUCUAGAGUGUUCCAAUCCCG ....(((....)))(((((.(.(((((((...((((((.(((....((---------------------(........-)))......))))))))).....((.(((((....))))).))..))))))).).))))). ( -32.80, z-score = -1.50, R) >consensus _UCUGGGAGUUCCUGGGAUCGGAACACUCAAGGAUUUCCUGUAAUCUUAAAAACGUUUAAAGUA___UGCAUACCUUC_GAGUUAAAGACAGAAAUUGAUUCGAAAGAACGAAAGUUUUCUCUAGAGUGUCCCAAUCCCG ....((.....)).(((((.((.((((((.((((((......))))))......................................................((.(((((....))))).))..)))))).)).))))). (-21.10 = -22.50 + 1.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:44 2011