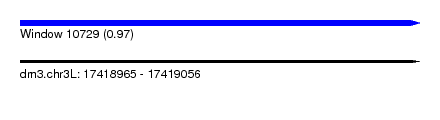
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,418,965 – 17,419,056 |
| Length | 91 |
| Max. P | 0.972083 |

| Location | 17,418,965 – 17,419,056 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 58.28 |
| Shannon entropy | 0.83407 |
| G+C content | 0.49034 |
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -12.44 |
| Energy contribution | -11.75 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.42 |
| Mean z-score | -0.54 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.972083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17418965 91 + 24543557 -------------UAAAACCAAUCACUUGAAUAUUCUUUCUUGUUCUCUCAUUUCAGCUGGCGCUUUGUCGCGUGCCGCUUACCA---CGGUGGACAUGGUCCCCAC -------------....((((.......(((......))).(((((.((......(((.(((((........)))))))).....---.)).)))))))))...... ( -19.90, z-score = -1.05, R) >droGri2.scaffold_15110 13217365 79 + 24565398 -------------------------UUAAUUAUGCUUAUGCAUCGUUUU---UGCAGCUGGUGCCAUGUCGCGUGCCGCUUACCAUAGCGGUGGACACCACCAUCAU -------------------------..(((.((((....)))).)))..---....(.(((((...((((.(..((((((......))))))))))).))))).).. ( -22.50, z-score = -0.61, R) >droVir3.scaffold_13049 19370450 86 - 25233164 -------------------AUUUAUUGACUUCAAAUUUGUGAUCCCUUU--CGGCAGCUGGUGCCAUGUCGCGUGCCGCCUACCACGGCGGUGGCGACCACAAGCAC -------------------.....(((....))).((((((........--.((((.....))))..(((((.(((((((......))))))))))))))))))... ( -30.60, z-score = -1.86, R) >droWil1.scaffold_180727 2491902 95 - 2741493 ---------UGAAAUCAAUUUUUACUGUAUUUCAUUUUGUUUUUUUUGUUCCUGUAGCUGGUGCCAUGUCUCGCGCCGCUUACCA---UGGUGGUCACCAUCAGCAU ---------((((((((........)).))))))......................(((((((....(.....)((((((.....---.))))))...))))))).. ( -19.10, z-score = -0.63, R) >droPer1.super_35 505195 88 + 973955 ------------------UUGGUAAUUCAUUA-AUGUGUAAAUCCUUGUAACUCCAGCUGGUGCCAUGUCGCGCGCUGCCUACCACGCUGGCGGACACCACCAGCAC ------------------..((.....(((..-..))).....))...........(((((((...((((.(((.(.((.......)).)))))))).))))))).. ( -25.30, z-score = -0.60, R) >dp4.chrXR_group6 4096573 88 - 13314419 ------------------UUGGUAAUUCAUUA-AUGUGUAAUUCCUUGUAAUUCCAGCUGGUGCCAUGUCGCGCGCUGCCUACCACGCUGGCGGACACCACCAGCAC ------------------..((.(((((((..-..))).))))))...........(((((((...((((.(((.(.((.......)).)))))))).))))))).. ( -26.50, z-score = -1.07, R) >droAna3.scaffold_13337 8371539 78 + 23293914 --------------------------ACAAGAAAUGACUAAUAAUCUCUAUUUGCAGUUGGCGCUCUGUCCCGUGCCGCUUACCA---CGGUGGCCACCACGAGCAC --------------------------...(((.((........)).)))....((.....))((((.((...((((((((.....---.))))).))).)))))).. ( -16.30, z-score = 0.46, R) >droEre2.scaffold_4784 9332475 101 - 25762168 AUGCGUUUCAUACUCACACAAUGCACUGAAAUAUUAUUAU---ACCUUUCAUUUCAGCUGGCGCUUUGUCGCGUGCCGCUUACCA---CGGUGGACAUGGUCCCCAC .((((((............))))))(((((((........---.......)))))))..(((((........)))))........---.((.((((...)))))).. ( -21.76, z-score = -0.26, R) >droYak2.chr3L 7550403 99 - 24197627 AAGAGUUUCAUAC--GCACCAUUCACGAAAAUAUUCUUGU---ACCUUUCAUUUUAGCUGGCGCCUUGUCGCGUGCCGCUUACCA---CGGUGGACAUGGUCCCCAC ..(.(..((((..--.((((......((((.(((....))---)..)))).....(((.(((((........)))))))).....---.))))...))))..).).. ( -19.50, z-score = 0.59, R) >droSec1.super_0 9471404 90 + 21120651 --------------AAAAUAAUUCACUUAGAUAUUCUUCCUUGAUCUUUCAUUUCAGCUGGCGCUUUGUCGCGUGCCGCUUACCA---CGGUGGACAUGGUCCCCAC --------------...............((...((......))....)).....(((.(((((........)))))))).....---.((.((((...)))))).. ( -18.00, z-score = -0.36, R) >droSim1.chr3L 16759906 91 + 22553184 -------------AAAAAAAAUUCACUUAAAUAUUAUUUCUUGAUCUUUAAUUUCAGCUGGCGCUUUGUCGCGUGCCGCUUACCA---CGGUGGCCAUGGUCCCCAC -------------.............(((((.((((.....)))).)))))....(((.(((((........)))))))).....---.((.(((....))).)).. ( -17.90, z-score = -0.57, R) >consensus _________________AUAAUUCACUUAAAUAAUCUUUUUUUACCUUUAAUUUCAGCUGGCGCCAUGUCGCGUGCCGCUUACCA___CGGUGGACACCAUCACCAC ........................................................((.(((((........)))))))(((((.....)))))............. (-12.44 = -11.75 + -0.69)

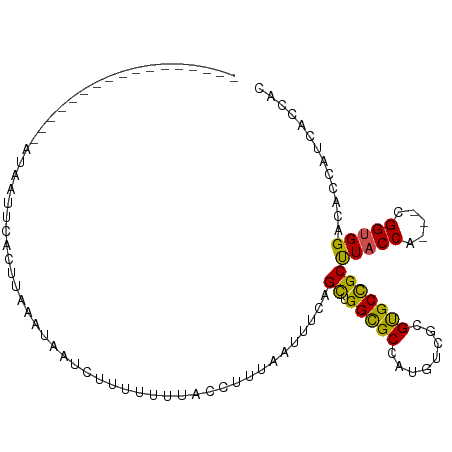
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:39 2011