| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,545,864 – 5,545,955 |
| Length | 91 |
| Max. P | 0.999741 |

| Location | 5,545,864 – 5,545,955 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 63.50 |
| Shannon entropy | 0.73634 |
| G+C content | 0.36712 |
| Mean single sequence MFE | -22.09 |
| Consensus MFE | -8.75 |
| Energy contribution | -7.71 |
| Covariance contribution | -1.04 |
| Combinations/Pair | 1.61 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.29 |
| SVM RNA-class probability | 0.999741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
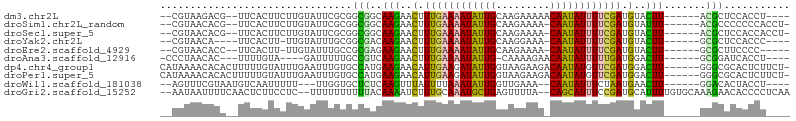
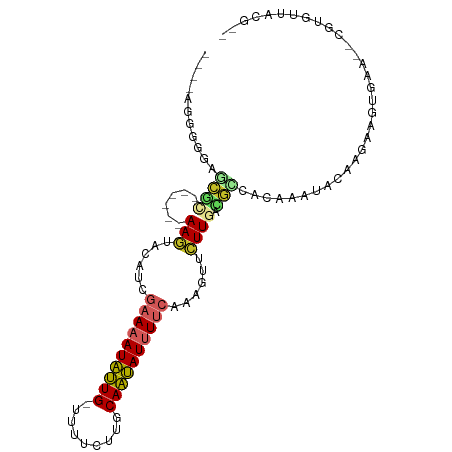
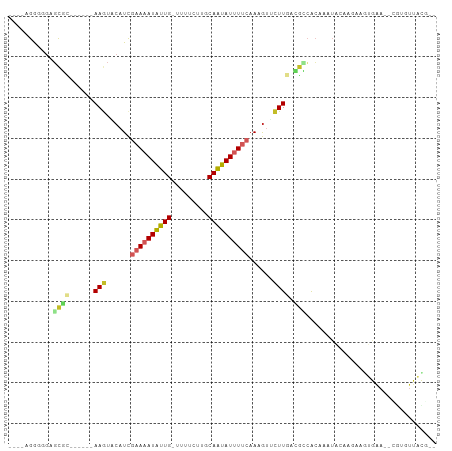
>dm3.chr2L 5545864 91 + 23011544 --CGUAAGACG--UUCACUUCUUGUAUUCGCGGCGGCAAGAACUUUGAAAAUAUUGCAAGAAAAACAAUAUUUUCGAUGUACUU------ACGCUCCACCU---- --(((((((((--((...(((((((..........)))))))....((((((((((.........))))))))))))))).)))------)))........---- ( -22.60, z-score = -2.59, R) >droSim1.chr2L_random 354958 93 + 909653 --CGUAACACG--UUCACUUCUUGUAUUCGCGGCGGCAAGAACUUUGAAAAUAUUGCAAGAAAA-CAAUAUUUUCGAUGUAUUU------ACGCCCCCCCACCU- --.........--..................((((..(((.((.((((((((((((........-)))))))))))).)).)))------.)))).........- ( -19.50, z-score = -1.90, R) >droSec1.super_5 3624313 93 + 5866729 --CGUAACACG--UUCACUUCUUGUAUUCGCGGCGGCAAGAACUUUGAAAAUAUUGCAAGAAAA-CAAUAUUUUCGAUGUACUU------ACGCUCCACCACCU- --.........--................(.((((..(((.((.((((((((((((........-)))))))))))).)).)))------.)))).).......- ( -20.20, z-score = -2.15, R) >droYak2.chr2L 8675655 87 - 22324452 --CGUAACA----UUCACUU-UUGUAUUUGCGGCGACAAGAACUUUGAAAAUAUUGCAAGGAAA-CAAUAUUUUCGAUGUACUU------GCGCUCCACCC---- --(((((((----.......-.)))...))))(((.((((.((.((((((((((((........-)))))))))))).)).)))------)))).......---- ( -24.30, z-score = -3.89, R) >droEre2.scaffold_4929 5632384 88 + 26641161 --CGUAACACC--UUCACUU-UUGUAUUUGCCGCGAGAAGAACUUUGAAAAUAUUGCAAGAAAA-CAAUAUUUUCGAUGUACUU------GCGCUUCCCC----- --.........--.......-...........(((..(((.((.((((((((((((........-)))))))))))).)).)))------.)))......----- ( -18.50, z-score = -2.77, R) >droAna3.scaffold_12916 4478215 86 + 16180835 -CCCUAACAC---UUUUGUA----GAUUUUUGCCGUCAAGAACUUUGAAAAUAUUG-CAAAAGAACAAUAUUUUUGAUGGACUU------GCGGAUCACCU---- -.........---.......----........(((.((((..(.(..(((((((((-........)))))))))..).)..)))------)))).......---- ( -18.70, z-score = -2.74, R) >dp4.chr4_group1 4863389 98 + 5278887 CAUAAAACACACUUUUUGUAUUUGAAUUUGUGCCAUGAAGAACAUUGAAGAUAUUGGUAAGAAGACAAUAUGUUCGAUGGACUU------GGGCGCACUCUUCU- .((((((......))))))....(((..((((((...(((..(((((((.((((((.........)))))).)))))))..)))------.))))))...))).- ( -27.20, z-score = -3.50, R) >droPer1.super_5 6460006 98 - 6813705 CAUAAAACACACUUUUUGUAUUUGAAUUUGUGCCAUGAAGAACAUUGAAGAUAUUGGUAAGAAGACAAUAUGUUCGAUGGACUU------GGGCGCACUCUUCU- .((((((......))))))....(((..((((((...(((..(((((((.((((((.........)))))).)))))))..)))------.))))))...))).- ( -27.20, z-score = -3.50, R) >droWil1.scaffold_181038 451503 88 + 637489 --AGUUUCGUAAUGUCAAUUUUU---UUGGUGCUCUCAAGUUUAUUUUAAAUAUUGGUUGAAA--CAAUAUUUCUAAUGAACUU------GGACACUACCU---- --.....................---.(((((.((.((((((((((..((((((((.......--))))))))..)))))))))------))))))))...---- ( -23.60, z-score = -4.56, R) >droGri2.scaffold_15252 6037194 99 - 17193109 --AAUAAUUUUCAACUCUUCCUC--UUUUUUUUUUACAAAAUCUUUGCAAAUGCUGAGUUUUA--CAGCAUUUCCGAUGCAUUUUGUGCAAAGAACACCCCUCAA --.....................--..((((((.(((((((((.(((.((((((((.......--)))))))).))).).)))))))).)))))).......... ( -19.10, z-score = -3.46, R) >consensus __CGUAACACA__UUCACUUCUUGUAUUUGCGGCGUCAAGAACUUUGAAAAUAUUGCAAGAAAA_CAAUAUUUUCGAUGUACUU______GCGCUCCACCU____ .............................((......(((..(.((((((((((((.........)))))))))))).)..)))........))........... ( -8.75 = -7.71 + -1.04)

| Location | 5,545,864 – 5,545,955 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 63.50 |
| Shannon entropy | 0.73634 |
| G+C content | 0.36712 |
| Mean single sequence MFE | -22.49 |
| Consensus MFE | -6.12 |
| Energy contribution | -6.36 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
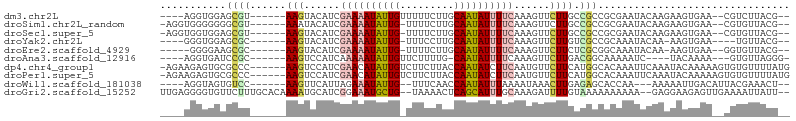
>dm3.chr2L 5545864 91 - 23011544 ----AGGUGGAGCGU------AAGUACAUCGAAAAUAUUGUUUUUCUUGCAAUAUUUUCAAAGUUCUUGCCGCCGCGAAUACAAGAAGUGAA--CGUCUUACG-- ----..((((.(.((------(((.((...(((((((((((.......)))))))))))...)).)))))).))))......((((.(....--).))))...-- ( -25.70, z-score = -2.63, R) >droSim1.chr2L_random 354958 93 - 909653 -AGGUGGGGGGGCGU------AAAUACAUCGAAAAUAUUG-UUUUCUUGCAAUAUUUUCAAAGUUCUUGCCGCCGCGAAUACAAGAAGUGAA--CGUGUUACG-- -.(((((((((((((------....))...((((((((((-(......)))))))))))...)))))).)))))(((..(((.....)))..--)))......-- ( -27.50, z-score = -2.43, R) >droSec1.super_5 3624313 93 - 5866729 -AGGUGGUGGAGCGU------AAGUACAUCGAAAAUAUUG-UUUUCUUGCAAUAUUUUCAAAGUUCUUGCCGCCGCGAAUACAAGAAGUGAA--CGUGUUACG-- -.(((((((((((((------....))...((((((((((-(......)))))))))))...)))).)))))))(((..(((.....)))..--)))......-- ( -28.70, z-score = -3.02, R) >droYak2.chr2L 8675655 87 + 22324452 ----GGGUGGAGCGC------AAGUACAUCGAAAAUAUUG-UUUCCUUGCAAUAUUUUCAAAGUUCUUGUCGCCGCAAAUACAA-AAGUGAA----UGUUACG-- ----..((((...((------(((.((...((((((((((-(......)))))))))))...)).)))))..))))........-..((((.----..)))).-- ( -23.40, z-score = -2.62, R) >droEre2.scaffold_4929 5632384 88 - 26641161 -----GGGGAAGCGC------AAGUACAUCGAAAAUAUUG-UUUUCUUGCAAUAUUUUCAAAGUUCUUCUCGCGGCAAAUACAA-AAGUGAA--GGUGUUACG-- -----((((((((..------..)).....((((((((((-(......)))))))))))....)))))).((..(((..(((..-..)))..--..)))..))-- ( -22.40, z-score = -2.71, R) >droAna3.scaffold_12916 4478215 86 - 16180835 ----AGGUGAUCCGC------AAGUCCAUCAAAAAUAUUGUUCUUUUG-CAAUAUUUUCAAAGUUCUUGACGGCAAAAAUC----UACAAAA---GUGUUAGGG- ----((((...((((------(((..(.(..((((((((((......)-)))))))))..).)..)))).))).....)))----)......---.........- ( -17.50, z-score = -1.82, R) >dp4.chr4_group1 4863389 98 - 5278887 -AGAAGAGUGCGCCC------AAGUCCAUCGAACAUAUUGUCUUCUUACCAAUAUCUUCAAUGUUCUUCAUGGCACAAAUUCAAAUACAAAAAGUGUGUUUUAUG -.(((...((.(((.------(((..(((.(((.((((((.........)))))).))).)))..)))...))).))..)))(((((((.....))))))).... ( -21.10, z-score = -2.96, R) >droPer1.super_5 6460006 98 + 6813705 -AGAAGAGUGCGCCC------AAGUCCAUCGAACAUAUUGUCUUCUUACCAAUAUCUUCAAUGUUCUUCAUGGCACAAAUUCAAAUACAAAAAGUGUGUUUUAUG -.(((...((.(((.------(((..(((.(((.((((((.........)))))).))).)))..)))...))).))..)))(((((((.....))))))).... ( -21.10, z-score = -2.96, R) >droWil1.scaffold_181038 451503 88 - 637489 ----AGGUAGUGUCC------AAGUUCAUUAGAAAUAUUG--UUUCAACCAAUAUUUAAAAUAAACUUGAGAGCACCAA---AAAAAUUGACAUUACGAAACU-- ----..(((((((((------(((((.(((..((((((((--.......))))))))..))).)))))).(.....)..---.......))))))))......-- ( -19.50, z-score = -3.96, R) >droGri2.scaffold_15252 6037194 99 + 17193109 UUGAGGGGUGUUCUUUGCACAAAAUGCAUCGGAAAUGCUG--UAAAACUCAGCAUUUGCAAAGAUUUUGUAAAAAAAAAA--GAGGAAGAGUUGAAAAUUAUU-- .....((.(.(((((((((.....)))...(.((((((((--.......)))))))).).....................--)))))).).))..........-- ( -18.00, z-score = -0.43, R) >consensus ____AGGGGGAGCGC______AAGUACAUCGAAAAUAUUG_UUUUCUUGCAAUAUUUUCAAAGUUCUUGACGCCACAAAUACAAGAAGUGAA__CGUGUUACG__ ..............................((((((((((.........)))))))))).............................................. ( -6.12 = -6.36 + 0.24)
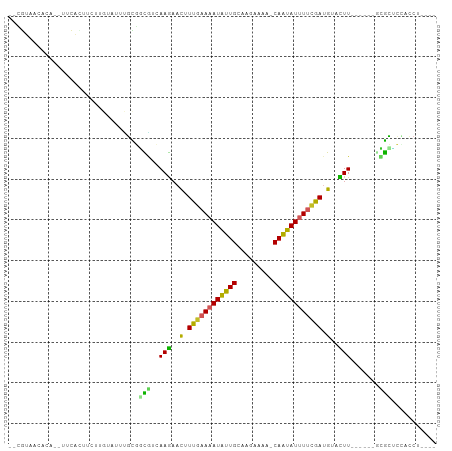
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:19 2011