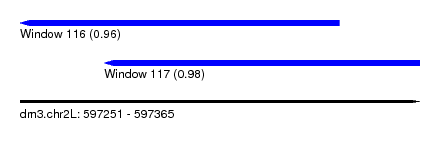
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 597,251 – 597,365 |
| Length | 114 |
| Max. P | 0.978022 |

| Location | 597,251 – 597,342 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.53 |
| Shannon entropy | 0.41471 |
| G+C content | 0.47733 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -13.41 |
| Energy contribution | -15.99 |
| Covariance contribution | 2.58 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.963710 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 597251 91 - 23011544 ---------UCAAUCGCAAAUCCUCAGC-ACGGGAUUUGGCUGCUGAAUCCGAAGAAUCUGAUUCC--GAUUCUGG--UGCGGAUUCUG-CUGCUGAUUUCGAUUU------ ---------..(((((((((((((....-..))))))))((.((.(((((((.((((((.......--))))))..--..))))))).)-).))......))))).------ ( -33.90, z-score = -3.18, R) >droSim1.chr2L 603662 91 - 22036055 ---------UCAAUCGCAAAUCCUCAGC-ACGGGAUUUGGCUGCUGAAUCCGAAGAAUCUGAUUCC--GAUUCUGG--UGCGGAUUCUG-CUGCUGAUUUCGAUUU------ ---------..(((((((((((((....-..))))))))((.((.(((((((.((((((.......--))))))..--..))))))).)-).))......))))).------ ( -33.90, z-score = -3.18, R) >droSec1.super_14 579929 91 - 2068291 ---------UCAAUCGCAAAUCCUCAGC-ACGGGAUUUGGCUGCUGAAUCCGAAGAAUCUGAUUCC--GAUUCUGG--UGCGGAUUCUG-CUGCUGAUUUCGAUUU------ ---------..(((((((((((((....-..))))))))((.((.(((((((.((((((.......--))))))..--..))))))).)-).))......))))).------ ( -33.90, z-score = -3.18, R) >droYak2.chr2L 585825 91 - 22324452 ---------UCAAUUGCAAAUCCUCAGC-ACGGGAUUUGGCUGCUGAAUCCGAAGAAUCUGAUUCC--GAUUCUGG--UGCGGAUUCUG-CUGCUGAUUUCGAUUU------ ---------..(((((((((((((....-..))))))))((.((.(((((((.((((((.......--))))))..--..))))))).)-).))......))))).------ ( -31.80, z-score = -2.74, R) >droEre2.scaffold_4929 652947 91 - 26641161 ---------UCAAUUGCAAAUCCUCAGC-ACGGGAUUUGGCUGCUGAAUCCGAAGAAUCUGAUUCC--GAUUCUGG--UGCGGAUUCUG-CUGCUGAUUUCGAUUU------ ---------..(((((((((((((....-..))))))))((.((.(((((((.((((((.......--))))))..--..))))))).)-).))......))))).------ ( -31.80, z-score = -2.74, R) >droAna3.scaffold_12916 1600193 98 - 16180835 CCAUCGCCGGCAGUUUCAAAUCCUCAUCUUCAGGAUUUAGCUCCU---------GAAUCUGAUUCC--GAUUCUGG--UGCGGAUUCUG-CUGCUGAUGCUGAUUUCGAUUU .(((((.((((((..(((((((((.......))))))).((.((.---------(((((.......--))))).))--.)).))..)))-))).)))))............. ( -33.40, z-score = -2.95, R) >droPer1.super_1 7815204 96 + 10282868 ---------CCCGCUCCCGCUCCUGCUCCUGCUCGUGGAAUCCUUGAAUCC---CAAUCUGAUUUCUGGGUUCCGGAUUCCGGGAGCUGAUUUCUGAUUUCGAUUUCU---- ---------...(((((((.(((.((....))....(((((((..(((((.---......)))))..))))))))))...))))))).((..((.......))..)).---- ( -33.70, z-score = -3.35, R) >consensus _________UCAAUUGCAAAUCCUCAGC_ACGGGAUUUGGCUGCUGAAUCCGAAGAAUCUGAUUCC__GAUUCUGG__UGCGGAUUCUG_CUGCUGAUUUCGAUUU______ ...........(((((((((((((.......))))))))((....(((((((.((((((.........))))))......))))))).....))......)))))....... (-13.41 = -15.99 + 2.58)

| Location | 597,275 – 597,365 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 81.03 |
| Shannon entropy | 0.36023 |
| G+C content | 0.54815 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -26.65 |
| Energy contribution | -27.52 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978022 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 597275 90 - 23011544 UGUCGGCAGCAGGCGACUCAGCCUCAAUCGCAAAUCCUCAGCACGGGAUUUGGCUGCUGAAUCCGAAGAAUCUGAUUCCGAUUCUGGUGC--- ..((((((((((((......))))......((((((((......))))))))))))))))..((..((((((.......))))))))...--- ( -34.10, z-score = -2.13, R) >droSim1.chr2L 603686 90 - 22036055 UGUCGGCAGCAGGCGACUCAGCCUCAAUCGCAAAUCCUCAGCACGGGAUUUGGCUGCUGAAUCCGAAGAAUCUGAUUCCGAUUCUGGUGC--- ..((((((((((((......))))......((((((((......))))))))))))))))..((..((((((.......))))))))...--- ( -34.10, z-score = -2.13, R) >droSec1.super_14 579953 90 - 2068291 UGUCGGCAGCAGGCGACUCAGCCUCAAUCGCAAAUCCUCAGCACGGGAUUUGGCUGCUGAAUCCGAAGAAUCUGAUUCCGAUUCUGGUGC--- ..((((((((((((......))))......((((((((......))))))))))))))))..((..((((((.......))))))))...--- ( -34.10, z-score = -2.13, R) >droYak2.chr2L 585849 90 - 22324452 UGUCGGCAGCAGGCGACUCAGCCUCAAUUGCAAAUCCUCAGCACGGGAUUUGGCUGCUGAAUCCGAAGAAUCUGAUUCCGAUUCUGGUGC--- ..((((((((((((......))))......((((((((......))))))))))))))))..((..((((((.......))))))))...--- ( -34.10, z-score = -2.22, R) >droEre2.scaffold_4929 652971 90 - 26641161 UGUCGGCAGCAGGCGACUCAGACUCAAUUGCAAAUCCUCAGCACGGGAUUUGGCUGCUGAAUCCGAAGAAUCUGAUUCCGAUUCUGGUGC--- ..((((((((((....))............((((((((......))))))))))))))))..((..((((((.......))))))))...--- ( -28.90, z-score = -0.94, R) >droPer1.super_1 7815238 85 + 10282868 UGUCAGUGGCAGGCUCUCCGGCUCCCGCUCCCGCUCCU--------GCUCCUGCUCGUGGAAUCCUUGAAUCCCAAUCUGAUUUCUGGGUUCC .(.(((..(((((......(((....)))......)))--------))..))))....(((((((..(((((.......)))))..))))))) ( -27.90, z-score = -2.16, R) >consensus UGUCGGCAGCAGGCGACUCAGCCUCAAUCGCAAAUCCUCAGCACGGGAUUUGGCUGCUGAAUCCGAAGAAUCUGAUUCCGAUUCUGGUGC___ ..((((((((((((......))))......((((((((......))))))))))))))))......((((((.......))))))........ (-26.65 = -27.52 + 0.87)
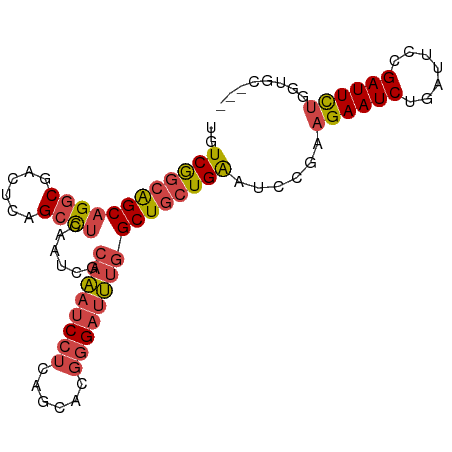
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:16 2011